Cell-type specific sequencing of microRNAs from complex animal tissues
- PMID: 29481550
- PMCID: PMC5886366
- DOI: 10.1038/nmeth.4610
Cell-type specific sequencing of microRNAs from complex animal tissues
Abstract
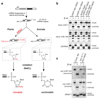
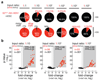
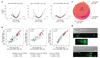
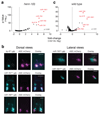
MicroRNAs (miRNAs) play an essential role in the post-transcriptional regulation of animal development and physiology. However, in vivo studies aimed at linking miRNA function to the biology of distinct cell types within complex tissues remain challenging, partly because in vivo miRNA-profiling methods lack cellular resolution. We report microRNome by methylation-dependent sequencing (mime-seq), an in vivo enzymatic small-RNA-tagging approach that enables high-throughput sequencing of tissue- and cell-type-specific miRNAs in animals. The method combines cell-type-specific 3'-terminal 2'-O-methylation of animal miRNAs by a genetically encoded, plant-specific methyltransferase (HEN1), with chemoselective small-RNA cloning and high-throughput sequencing. We show that mime-seq uncovers the miRNomes of specific cells within Caenorhabditis elegans and Drosophila at unprecedented specificity and sensitivity, enabling miRNA profiling with single-cell resolution in whole animals. Mime-seq overcomes current challenges in cell-type-specific small-RNA profiling and provides novel entry points for understanding the function of miRNAs in spatially restricted physiological settings.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

