Distinct fermentation and antibiotic sensitivity profiles exist in salmonellae of canine and human origin
- PMID: 29482521
- PMCID: PMC5828451
- DOI: 10.1186/s12866-018-1153-4
Distinct fermentation and antibiotic sensitivity profiles exist in salmonellae of canine and human origin
Abstract
Background: Salmonella enterica is a recognised cause of diarrhoea in dogs and humans, yet the potential for transfer of salmonellosis between dogs and their owners is unclear, with reported evidence both for and against Salmonella as a zoonotic pathogen. A collection of 174 S. enterica isolates from clinical infections in humans and dogs were analysed for serotype distribution, carbon source utilisation, chemical and antimicrobial sensitivity profiles. The aim of the study was to understand the degree of conservation in phenotypic characteristics of isolates across host species.
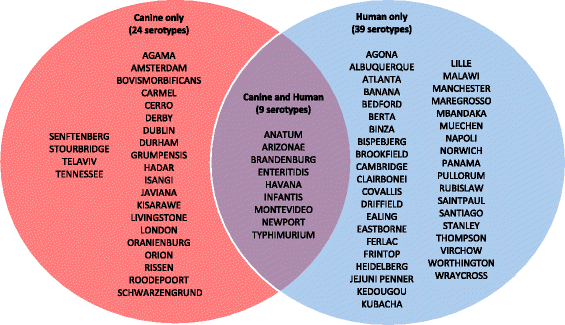
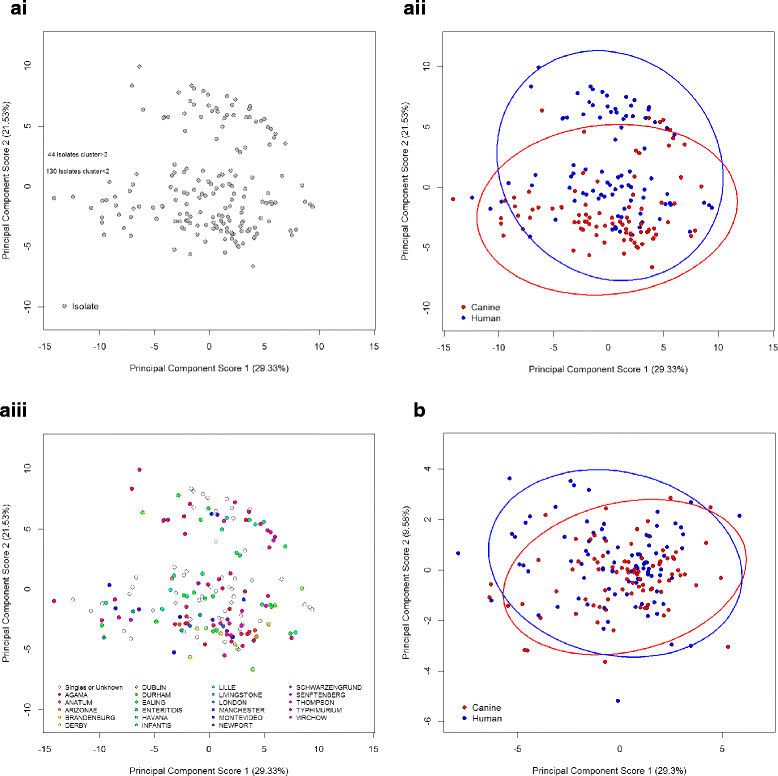
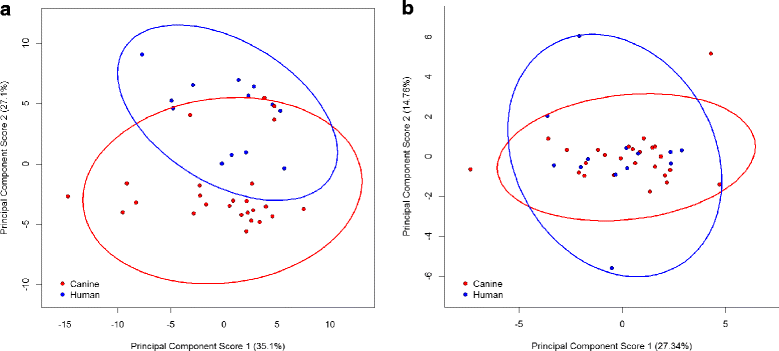
Results: Serovar distribution across human and canine isolates demonstrated nine serovars common to both host species, 24 serovars present in only the canine collection and 39 solely represented within the human collection. Significant differences in carbon source utilisation profiles and ampicillin, amoxicillin and chloramphenicol sensitivity profiles were detected in isolates of human and canine origin. Differences between the human and canine Salmonella collections were suggestive of evolutionary separation, with canine isolates better able to utilise several simple sugars than their human counterparts. Generally higher minimum inhibitory concentrations of three broad-spectrum antimicrobials, commonly used in veterinary medicine, were also observed in canine S. enterica isolates.
Conclusions: Differential carbon source utilisation and antimicrobial sensitivity profiles in pathogenic Salmonella isolated from humans and dogs are suggestive of distinct reservoirs of infection for these hosts. Although these findings do not preclude zoonotic or anthroponotic potential in salmonellae, the separation of carbon utilisation and antibiotic profiles with isolate source is indicative that infectious isolates are not part of a common reservoir shared frequently between these host species.
Keywords: Anthroponosis; Biolog; Dog; MIC; S. Enterica; Serovar; Zoonosis.
Conflict of interest statement
Ethics approval and consent to participate
Approval to conduct the collections and analysis of salmonellae from canine faeces was granted by the WALTHAM Centre for Pet Nutrition Animal Welfare and Ethical Review Board. The use of archived
Consent for publication
Not applicable.
Competing interests
This research was funded through a Biotechnology & Biological Sciences Research Council (BBSRC) CASE studentship granted to Aston University and undertaken in collaboration with the WALTHAM Centre for Pet Nutrition (Mars Petcare). The BBSRC funded Aston University researchers Professor Anthony Hilton and the recipient of the BBSRC studentship, Preena Lowden. Corrin Wallis and Zoe Marshall-Jones represented industrial partner supervisors employed by Mars Petcare UK. Mars Petcare representatives were involved in the design of the study, data analysis and interpretation as well as in writing the manuscript. There are no products in development or marketed products to declare.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
Similar articles
-
Serovars, bacteriophage types and antimicrobial sensitivities associated with salmonellosis in dogs in the UK (1954-2012).Vet Rec. 2014 Jan 25;174(4):94. doi: 10.1136/vr.101864. Epub 2013 Nov 25. Vet Rec. 2014. PMID: 24277916
-
Drug resistance patterns of Salmonella isolates of equine origin from India.J Infect Dev Ctries. 2009 Mar 1;3(2):141-7. doi: 10.3855/jidc.61. J Infect Dev Ctries. 2009. PMID: 19755745
-
Virulotyping of Salmonella enterica serovar Napoli strains isolated in Italy from human and nonhuman sources.Foodborne Pathog Dis. 2011 Sep;8(9):997-1003. doi: 10.1089/fpd.2010.0833. Epub 2011 May 11. Foodborne Pathog Dis. 2011. PMID: 21561382
-
Salmonella genomic islands and antibiotic resistance in Salmonella enterica.Future Microbiol. 2010 Oct;5(10):1525-38. doi: 10.2217/fmb.10.122. Future Microbiol. 2010. PMID: 21073312 Review.
-
Molecular insights into farm animal and zoonotic Salmonella infections.Philos Trans R Soc Lond B Biol Sci. 2009 Sep 27;364(1530):2709-23. doi: 10.1098/rstb.2009.0094. Philos Trans R Soc Lond B Biol Sci. 2009. PMID: 19687040 Free PMC article. Review.
Cited by
-
Endogenous CRISPR-assisted microhomology-mediated end joining enables rapid genome editing in Zymomonas mobilis.Biotechnol Biofuels. 2021 Oct 24;14(1):208. doi: 10.1186/s13068-021-02056-z. Biotechnol Biofuels. 2021. PMID: 34689795 Free PMC article.
References
-
- Samadpour M, Barbour MW, Nguyen T, Cao TM, Buck F, Depavia GA, et al. Incidence of enterohemorrhagic Escherichia coli, Escherichia coli O157, Salmonella, and Listeria monocytogenes in retail fresh ground beef, sprouts, and mushrooms. J Food Prot. 2006;69(2):441–443. doi: 10.4315/0362-028X-69.2.441. - DOI - PubMed
-
- Regunath H, Salza W. Sources and vehicles of foodborne infectious diseases. In: Khardori N, editor. Food microbiology: in human health and disease FL. USA: CRC Press; 2016. pp. 67–91.
-
- Fardsanei F, Nikkhahi F, Bakhshi B, Salehi TZ, Tamai IA, Soltan Dallal MM. Molecular characterization of Salmonella enterica serotype Enteritidis isolates from food and human samples by serotyping, antimicrobial resistance, plasmid profiling, (GTG)5-PCR and ERIC-PCR. New Microbes New Infect. 2016;14:24–30. doi: 10.1016/j.nmni.2016.07.016. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

