Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease
- PMID: 29482626
- PMCID: PMC5827984
- DOI: 10.1186/s13072-018-0178-0
Alterations in sperm DNA methylation, non-coding RNA and histone retention associate with DDT-induced epigenetic transgenerational inheritance of disease
Abstract
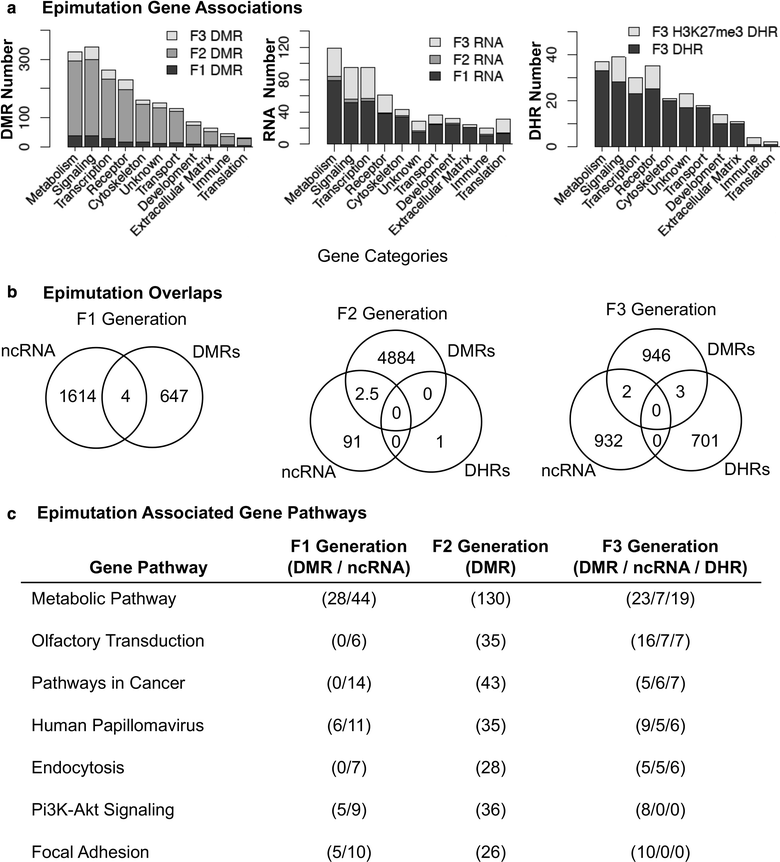
Background: Environmental toxicants such as DDT have been shown to induce the epigenetic transgenerational inheritance of disease (e.g., obesity) through the germline. The current study was designed to investigate the DDT-induced concurrent alterations of a number of different epigenetic processes including DNA methylation, non-coding RNA (ncRNA) and histone retention in sperm.
Methods: Gestating females were exposed transiently to DDT during fetal gonadal development, and then, the directly exposed F1 generation, the directly exposed germline F2 generation and the transgenerational F3 generation sperm were investigated.
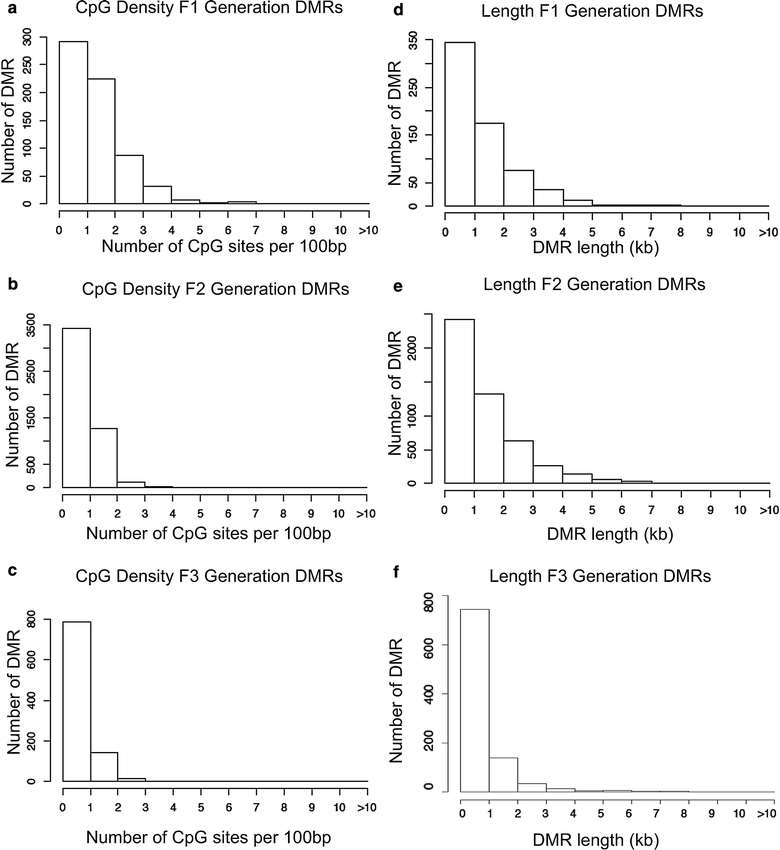
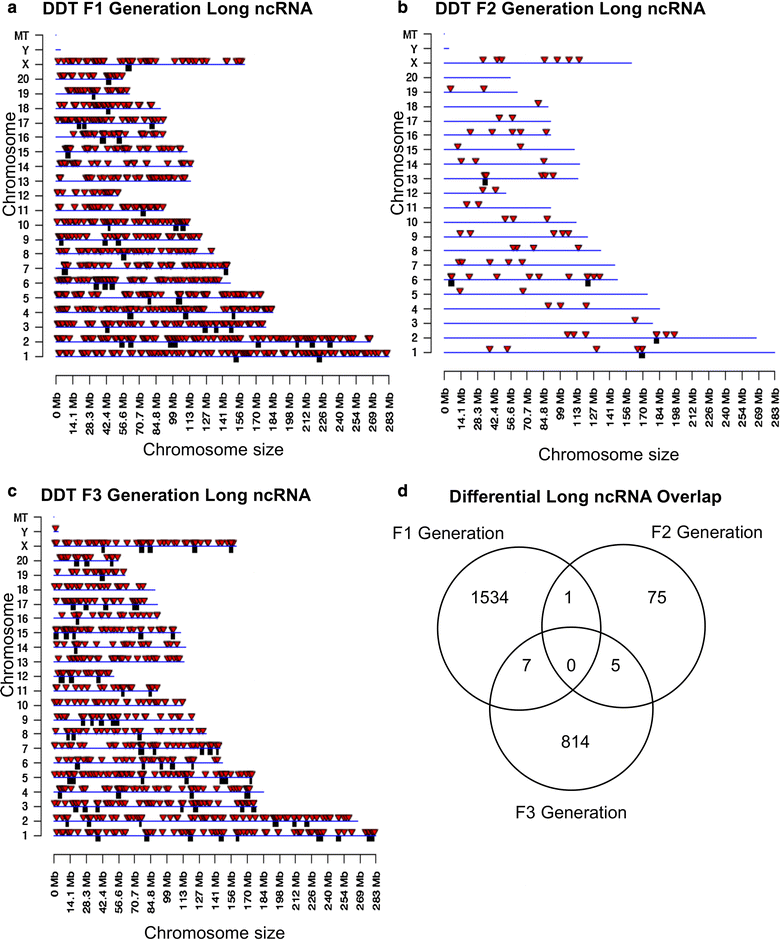
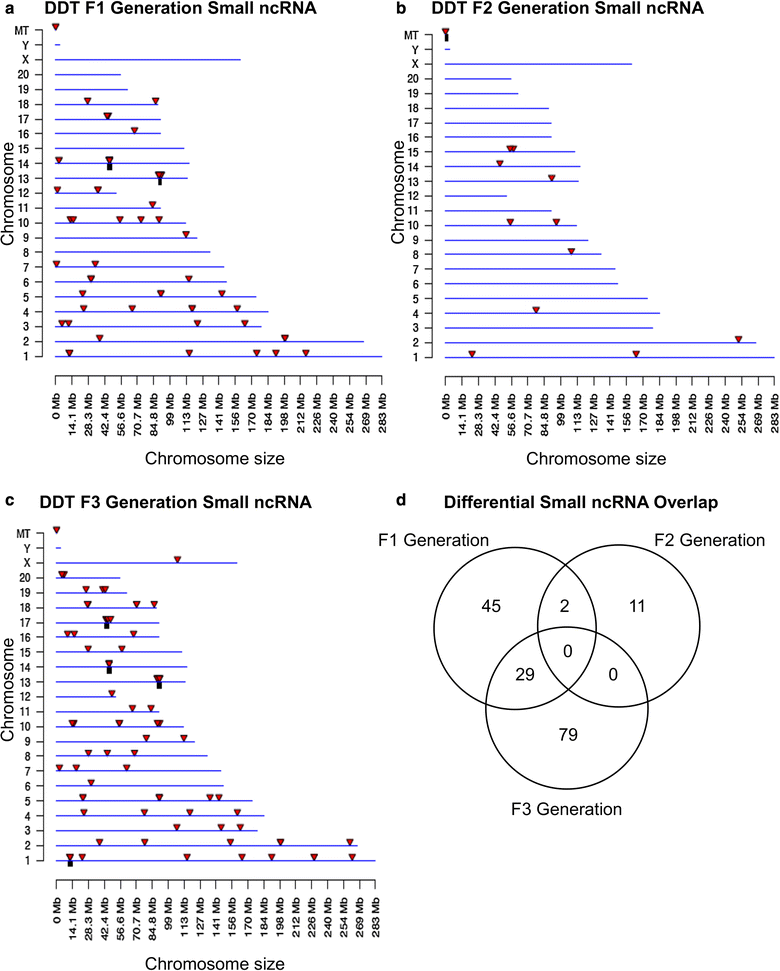
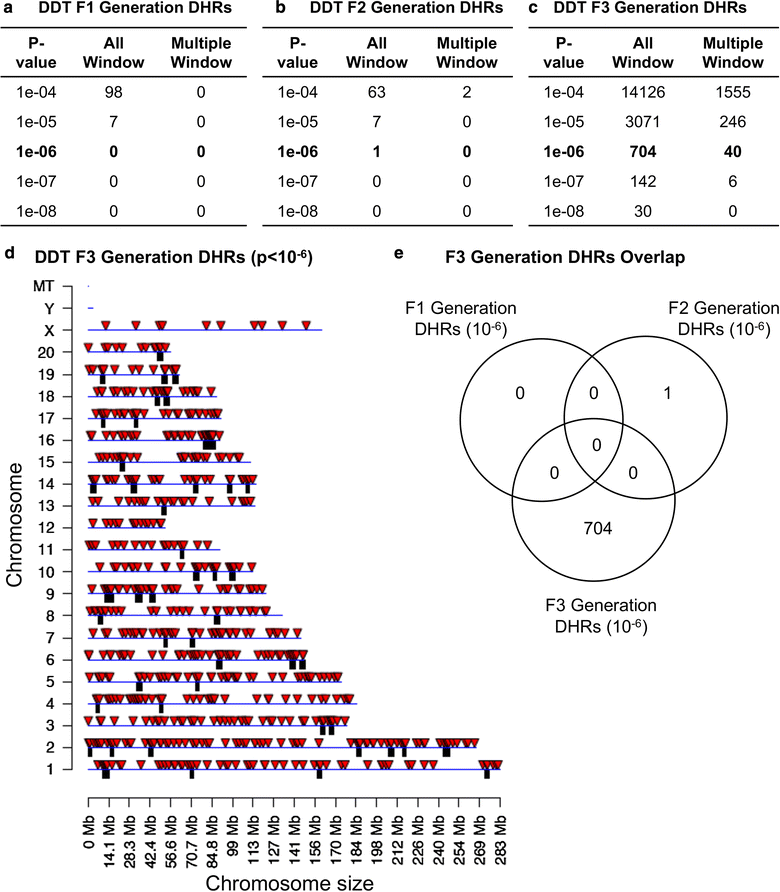
Results: DNA methylation and ncRNA were altered in each generation sperm with the direct exposure F1 and F2 generations being predominantly distinct from the F3 generation epimutations. The piRNA and small tRNA were the most predominant classes of ncRNA altered. A highly conserved set of histone retention sites were found in the control lineage generations which was not significantly altered between generations, but a large number of new histone retention sites were found only in the transgenerational generation DDT lineage sperm.
Conclusions: Therefore, all three different epigenetic processes were concurrently altered as DDT induced the epigenetic transgenerational inheritance of sperm epimutations. The direct exposure generations sperm epigenetic alterations were distinct from the transgenerational sperm epimutations. The genomic features and gene associations with the epimutations were investigated to help elucidate the integration of these different epigenetic processes. Observations demonstrate all three epigenetic processes are involved in transgenerational inheritance. The different epigenetic processes appear to be integrated in mediating the epigenetic transgenerational inheritance phenomenon.
Keywords: DDT; DNA methylation; Disease etiology; Epigenetic; Histone; Inheritance; Sperm; Transgenerational; ncRNA.
Figures





References
-
- McCreary JK, Truica LS, Friesen B, Yao Y, Olson DM, Kovalchuk I, Cross AR, Metz GA. Altered brain morphology and functional connectivity reflect a vulnerable affective state after cumulative multigenerational stress in rats. Neuroscience. 2016;330:79–89. doi: 10.1016/j.neuroscience.2016.05.046. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

