Improving saliva shotgun metagenomics by chemical host DNA depletion
- PMID: 29482639
- PMCID: PMC5827986
- DOI: 10.1186/s40168-018-0426-3
Improving saliva shotgun metagenomics by chemical host DNA depletion
Abstract
Background: Shotgun sequencing of microbial communities provides in-depth knowledge of the microbiome by cataloging bacterial, fungal, and viral gene content within a sample, providing an advantage over amplicon sequencing approaches that assess taxonomy but not function and are taxonomically limited. However, mammalian DNA can dominate host-derived samples, obscuring changes in microbial populations because few DNA sequence reads are from the microbial component. We developed and optimized a novel method for enriching microbial DNA from human oral samples and compared its efficiency and potential taxonomic bias with commercially available kits.
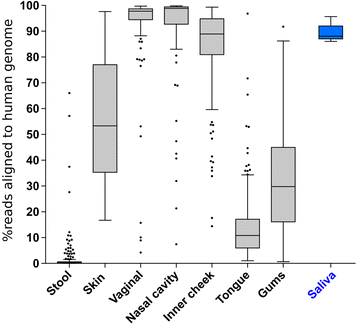
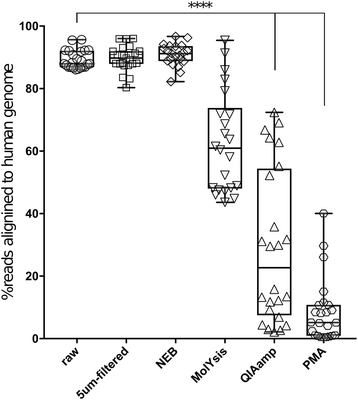
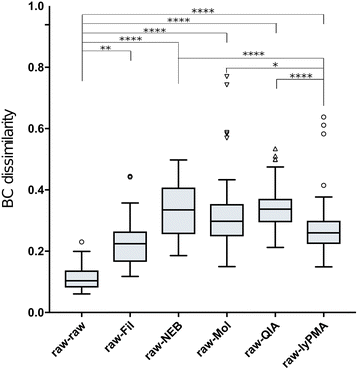
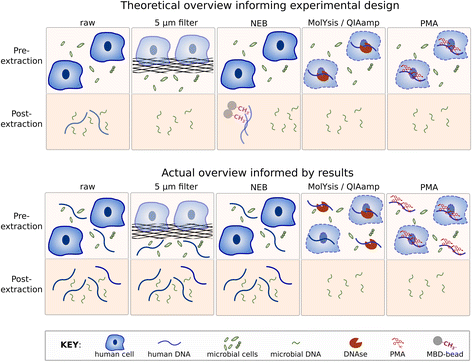
Results: Three commercially available host depletion kits were directly compared with size filtration and a novel method involving osmotic lysis and treatment with propidium monoazide (lyPMA) in human saliva samples. We evaluated the percentage of shotgun metagenomic sequencing reads aligning to the human genome, and taxonomic biases of those not aligning, compared to untreated samples. lyPMA was the most efficient method of removing host-derived sequencing reads compared to untreated sample (8.53 ± 0.10% versus 89.29 ± 0.03%). Furthermore, lyPMA-treated samples exhibit the lowest taxonomic bias compared to untreated samples.
Conclusion: Osmotic lysis followed by PMA treatment is a cost-effective, rapid, and robust method for enriching microbial sequence data in shotgun metagenomics from fresh and frozen saliva samples and may be extensible to other host-derived sample types.
Keywords: Host depletion; Microbial enrichment; Microbiome; Propidium monoazide; Saliva; Shotgun sequencing.
Conflict of interest statement
Ethics approval and consent to participate
All human subjects who participated in this project were consented under the UCSD HRPP approved protocol #150275 Explaining Variability in Human-associated microbial Communities PI Rob Knight (Federalwide Assurance number, FWA00004495).
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

