Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem
- PMID: 29482646
- PMCID: PMC5828423
- DOI: 10.1186/s40168-018-0420-9
Correcting for 16S rRNA gene copy numbers in microbiome surveys remains an unsolved problem
Abstract
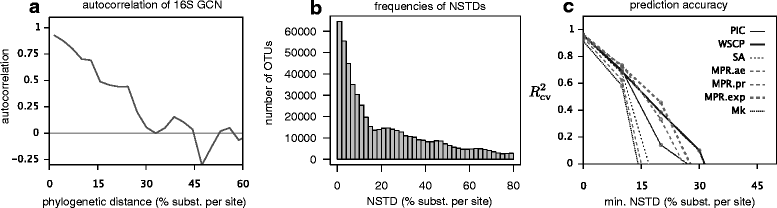
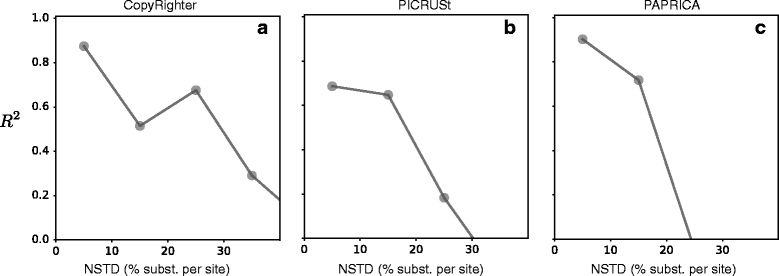
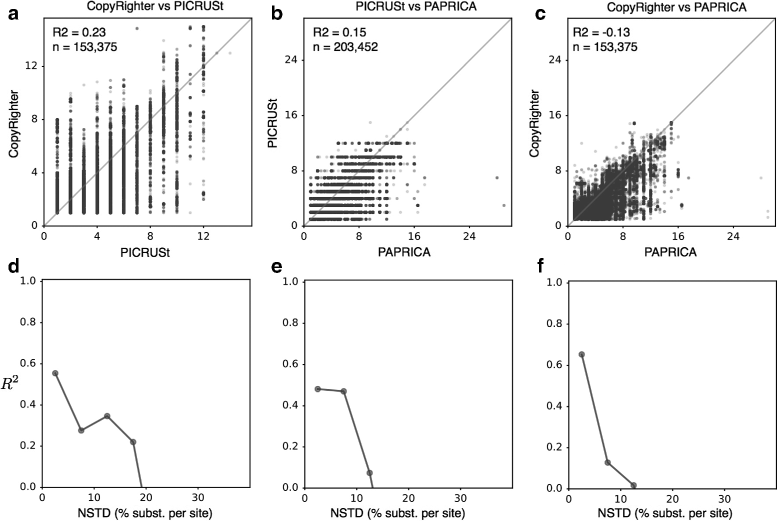
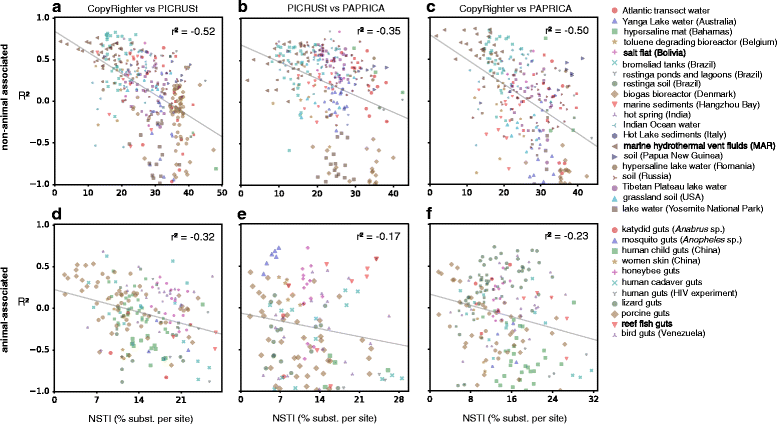
The 16S ribosomal RNA gene is the most widely used marker gene in microbial ecology. Counts of 16S sequence variants, often in PCR amplicons, are used to estimate proportions of bacterial and archaeal taxa in microbial communities. Because different organisms contain different 16S gene copy numbers (GCNs), sequence variant counts are biased towards clades with greater GCNs. Several tools have recently been developed for predicting GCNs using phylogenetic methods and based on sequenced genomes, in order to correct for these biases. However, the accuracy of those predictions has not been independently assessed. Here, we systematically evaluate the predictability of 16S GCNs across bacterial and archaeal clades, based on ∼ 6,800 public sequenced genomes and using several phylogenetic methods. Further, we assess the accuracy of GCNs predicted by three recently published tools (PICRUSt, CopyRighter, and PAPRICA) over a wide range of taxa and for 635 microbial communities from varied environments. We find that regardless of the phylogenetic method tested, 16S GCNs could only be accurately predicted for a limited fraction of taxa, namely taxa with closely to moderately related representatives (≲15% divergence in the 16S rRNA gene). Consistent with this observation, we find that all considered tools exhibit low predictive accuracy when evaluated against completely sequenced genomes, in some cases explaining less than 10% of the variance. Substantial disagreement was also observed between tools (R2<0.5) for the majority of tested microbial communities. The nearest sequenced taxon index (NSTI) of microbial communities, i.e., the average distance to a sequenced genome, was a strong predictor for the agreement between GCN prediction tools on non-animal-associated samples, but only a moderate predictor for animal-associated samples. We recommend against correcting for 16S GCNs in microbiome surveys by default, unless OTUs are sufficiently closely related to sequenced genomes or unless a need for true OTU proportions warrants the additional noise introduced, so that community profiles remain interpretable and comparable between studies.
Keywords: 16S rRNA; Gene copy number; Microbiome surveys; Phylogenetic reconstruction.
Conflict of interest statement
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Figures
References
-
- Lima-Mendez G, Faust K, Henry N, Decelle J, Colin S, Carcillo F, Chaffron S, Ignacio-Espinosa JC, Roux S, Vincent F, Bittner L, Darzi Y, Wang J, Audic S, Berline L, Bontempi G, Cabello AM, Coppola L, Cornejo-Castillo FM, d’Ovidio F, De Meester L, Ferrera I, Garet-Delmas MJ, Guidi L, Lara E, Pesant S, Royo-Llonch M, Salazar G, Sánchez P, Sebastian M, Souffreau C, Dimier C, Picheral M, Searson S, Kandels-Lewis S, coordinators TO, Gorsky G, Not F, Ogata H, Speich S, Stemmann L, Weissenbach J, Wincker P, Acinas SG, Sunagawa S, Bork P, Sullivan MB, Karsenti E, Bowler C, de Vargas C, Raes J. Determinants of community structure in the global plankton interactome. Science. 2015;348(6237):1262073. doi: 10.1126/science.1262073. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

