SARS-CoV related Betacoronavirus and diverse Alphacoronavirus members found in western old-world
- PMID: 29482919
- PMCID: PMC7112086
- DOI: 10.1016/j.virol.2018.01.014
SARS-CoV related Betacoronavirus and diverse Alphacoronavirus members found in western old-world
Abstract
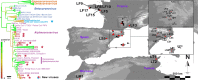
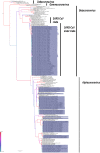
The emergence of SARS-CoV and MERS-CoV, triggered the discovery of a high diversity of coronaviruses in bats. Studies from Europe have shown that coronaviruses circulate in bats in France but this reflects only a fraction of the whole diversity. In the current study the diversity of coronaviruses circulating in western Europe was extensively explored. Ten alphacoronaviruses in eleven bat species belonging to the Miniopteridae, Vespertilionidae and Rhinolophidae families and, a SARS-CoV-related Betacoronavirus in Rhinolophus ferrumequinum were identified. The diversity and prevalence of bat coronaviruses presently reported from western Europe is much higher than previously described and includes a SARS-CoV sister group. This diversity demonstrates the dynamic evolution and circulation of coronaviruses in this species. That said, the identified coronaviruses were consistently associated with a particular bat species or genus, and these relationships were maintained no matter the geographic location. The observed phylogenetic grouping of coronaviruses from the same species in Europe and Asia, emphasizes the role of host/pathogen coevolution in this group.
Keywords: Bats; Chiroptera; Coronavirus; Diversity; Emergence; Europe; Evolution; MERS-CoV; Phylogenetics; SARS-CoV.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures

References
-
- Balboni A., Battilani M., Prosperi S. The SARS-like coronaviruses: the role of bats and evolutionary relationships with SARS coronavirus. New Microbiol. 2012;35:1–16. - PubMed
-
- Barataud M., Aulagnier S. Pourquoi certaines espèces de chauves-souris s'associent-elles en essaims mixtes durant lamise-bas et l'élevage des jeunes? Exemple en Limousin. Arvicola. 2012;20:40–42.
-
- Bermingham A., Chand M.A., Brown C.S., Aarons E., Tong C., Langrish C. Severe respiratory illness caused by a novel coronavirus, in a patient transferred to the United Kingdom from the middle East, September 2012. Eur. Surveill. Bull. Eur. Sur Mal. Transm. Eur. Commun. Dis. Bull. 2012;17:20290. - PubMed
-
- Crucitti P. Caratteristiche della aggregazione Miniopterus schreibersi - Myotis capaccinii nel Lazio, Italia centrale (Chiroptera) Boll. Mus. Reg. Sci. Nat. 1993;11:407–422.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

