ApoE facilitates the microglial response to amyloid plaque pathology
- PMID: 29483128
- PMCID: PMC5881464
- DOI: 10.1084/jem.20171265
ApoE facilitates the microglial response to amyloid plaque pathology
Abstract
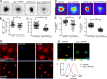
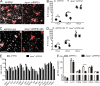
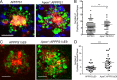
One of the hallmarks of Alzheimer's disease is the presence of extracellular diffuse and fibrillar plaques predominantly consisting of the amyloid-β (Aβ) peptide. Apolipoprotein E (ApoE) influences the deposition of amyloid pathology through affecting the clearance and aggregation of monomeric Aβ in the brain. In addition to influencing Aβ metabolism, increasing evidence suggests that apoE influences microglial function in neurodegenerative diseases. Here, we characterize the impact that apoE has on amyloid pathology and the innate immune response in APPPS1ΔE9 and APPPS1-21 transgenic mice. We report that Apoe deficiency reduced fibrillar plaque deposition, consistent with previous studies. However, fibrillar plaques in Apoe-deficient mice exhibited a striking reduction in plaque compaction. Hyperspectral fluorescent imaging using luminescent conjugated oligothiophenes identified distinct Aβ morphotypes in Apoe-deficient mice. We also observed a significant reduction in fibrillar plaque-associated microgliosis and activated microglial gene expression in Apoe-deficient mice, along with significant increases in dystrophic neurites around fibrillar plaques. Our results suggest that apoE is critical in stimulating the innate immune response to amyloid pathology.
© 2018 Ulrich et al.
Figures

References
-
- Bales K.R., Verina T., Cummins D.J., Du Y., Dodel R.C., Saura J., Fishman C.E., DeLong C.A., Piccardo P., Petegnief V., et al. . 1999. Apolipoprotein E is essential for amyloid deposition in the APP(V717F) transgenic mouse model of Alzheimer’s disease. Proc. Natl. Acad. Sci. USA. 96:15233–15238. 10.1073/pnas.96.26.15233 - DOI - PMC - PubMed
-
- Bell R.D., Sagare A.P., Friedman A.E., Bedi G.S., Holtzman D.M., Deane R., and Zlokovic B.V.. 2007. Transport pathways for clearance of human Alzheimer’s amyloid β-peptide and apolipoproteins E and J in the mouse central nervous system. J. Cereb. Blood Flow Metab. 27:909–918. 10.1038/sj.jcbfm.9600419 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

