Forecasting the spatial transmission of influenza in the United States
- PMID: 29483256
- PMCID: PMC5856508
- DOI: 10.1073/pnas.1708856115
Forecasting the spatial transmission of influenza in the United States
Abstract
Recurrent outbreaks of seasonal and pandemic influenza create a need for forecasts of the geographic spread of this pathogen. Although it is well established that the spatial progression of infection is largely attributable to human mobility, difficulty obtaining real-time information on human movement has limited its incorporation into existing infectious disease forecasting techniques. In this study, we develop and validate an ensemble forecast system for predicting the spatiotemporal spread of influenza that uses readily accessible human mobility data and a metapopulation model. In retrospective state-level forecasts for 35 US states, the system accurately predicts local influenza outbreak onset,-i.e., spatial spread, defined as the week that local incidence increases above a baseline threshold-up to 6 wk in advance of this event. In addition, the metapopulation prediction system forecasts influenza outbreak onset, peak timing, and peak intensity more accurately than isolated location-specific forecasts. The proposed framework could be applied to emergent respiratory viruses and, with appropriate modifications, other infectious diseases.
Keywords: data assimilation; human mobility; influenza forecast; metapopulation model; spatial transmission.
Conflict of interest statement
Conflict of interest statement: J.S. discloses partial ownership of SK Analytics. S.P., S.K., and W.Y. disclose consultation for SK Analytics.
Figures
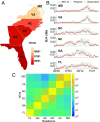
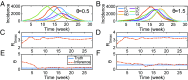
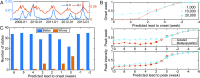
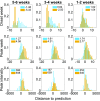
References
-
- Viboud C, et al. Synchrony, waves, and spatial hierarchies in the spread of influenza. Science. 2006;312:447–451. - PubMed
Publication types
MeSH terms
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

