RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome
- PMID: 29483269
- PMCID: PMC5856561
- DOI: 10.1073/pnas.1800038115
RNA self-assembly contributes to stress granule formation and defining the stress granule transcriptome
Abstract
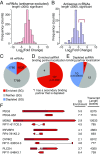
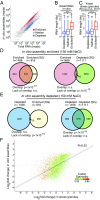
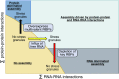
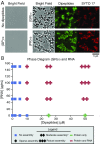
Stress granules are higher order assemblies of nontranslating mRNAs and proteins that form when translation initiation is inhibited. Stress granules are thought to form by protein-protein interactions of RNA-binding proteins. We demonstrate RNA homopolymers or purified cellular RNA forms assemblies in vitro analogous to stress granules. Remarkably, under conditions representative of an intracellular stress response, the mRNAs enriched in assemblies from total yeast RNA largely recapitulate the stress granule transcriptome. We suggest stress granules are formed by a summation of protein-protein and RNA-RNA interactions, with RNA self-assembly likely to contribute to other RNP assemblies wherever there is a high local concentration of RNA. RNA assembly in vitro is also increased by GR and PR dipeptide repeats, which are known to increase stress granule formation in cells. Since GR and PR dipeptides are involved in neurodegenerative diseases, this suggests that perturbations increasing RNA-RNA assembly in cells could lead to disease.
Keywords: RNA self-assembly; RNP granules; dipeptides; stress granules.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

