Network Motifs Capable of Decoding Transcription Factor Dynamics
- PMID: 29483553
- PMCID: PMC5827039
- DOI: 10.1038/s41598-018-21945-2
Network Motifs Capable of Decoding Transcription Factor Dynamics
Abstract
Transcription factors (TFs) can encode the information of upstream signal in terms of its temporal activation dynamics. However, it remains unclear how different types of TF dynamics are decoded by downstream signalling networks. In this work, we studied all three-node transcriptional networks for their ability to distinguish two types of TF dynamics: amplitude modulation (AM), where the TF is activated with a constant amplitude, and frequency modulation (FM), where the TF activity displays an oscillatory behavior. We found two sets of network topologies: one set can differentially respond to AM TF signal but not to FM; the other set to FM signal but not to AM. Interestingly, there is little overlap between the two sets. We identified the prevalent topological features in each set and gave a mechanistic explanation as to why they can differentially respond to only one type of TF signal. We also found that some network topologies have a weak (not robust) ability to differentially respond to both AM and FM input signals by using different values of parameters for AM and FM cases. Our results provide a novel network mechanism for decoding different TF dynamics.
Conflict of interest statement
The authors declare no competing interests.
Figures

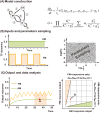

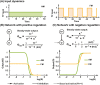

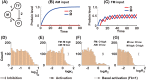

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

