HIF1α-dependent glycolysis promotes macrophage functional activities in protecting against bacterial and fungal infection
- PMID: 29483608
- PMCID: PMC5827022
- DOI: 10.1038/s41598-018-22039-9
HIF1α-dependent glycolysis promotes macrophage functional activities in protecting against bacterial and fungal infection
Abstract
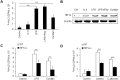
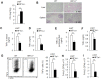
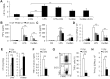
Macrophages are important innate immune defense system cells in the fight against bacterial and fungal pathogenic infections. They exhibit significant plasticity, particularly with their ability to undergo functional differentiation. Additionally, HIF1α is critically involved in the functional differentiation of macrophages during inflammation. However, the role of macrophage HIF1α in protecting against different pathogenic infections remains unclear. In this study, we investigated and compared the roles of HIF1α in different macrophage functional effects of bacterial and fungal infections in vitro and in vivo. We found that bacterial and fungal infections produced similar effects on macrophage functional differentiation. HIF1α deficiency inhibited pro-inflammatory macrophage functional activities when cells were stimulated with LPS or curdlan in vitro or when mice were infected with L. monocytogenes or C. albicans in vivo, thus decreasing pro-inflammatory TNFα and IL-6 secretion associated with pathogenic microorganism survival. Alteration of glycolytic pathway activation was required for the functional differentiation of pro-inflammatory macrophages in protecting against bacterial and fungal infections. Thus, the HIF1α-dependent glycolytic pathway is essential for pro-inflammatory macrophage functional differentiation in protecting against bacterial and fungal infections.
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Murray, P. J. Macrophage Polarization. Annual review of physiology (2016). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

