A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters
- PMID: 29483642
- PMCID: PMC5866759
- DOI: 10.1038/s41589-018-0004-9
A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters
Erratum in
-
Publisher Correction: A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters.Nat Chem Biol. 2018 Sep;14(9):901. doi: 10.1038/s41589-018-0023-6. Nat Chem Biol. 2018. PMID: 29520042
Abstract
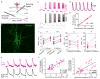
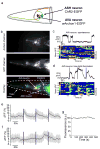
We developed a new way to engineer complex proteins toward multidimensional specifications using a simple, yet scalable, directed evolution strategy. By robotically picking mammalian cells that were identified, under a microscope, as expressing proteins that simultaneously exhibit several specific properties, we can screen hundreds of thousands of proteins in a library in just a few hours, evaluating each along multiple performance axes. To demonstrate the power of this approach, we created a genetically encoded fluorescent voltage indicator, simultaneously optimizing its brightness and membrane localization using our microscopy-guided cell-picking strategy. We produced the high-performance opsin-based fluorescent voltage reporter Archon1 and demonstrated its utility by imaging spiking and millivolt-scale subthreshold and synaptic activity in acute mouse brain slices and in larval zebrafish in vivo. We also measured postsynaptic responses downstream of optogenetically controlled neurons in C. elegans.
Conflict of interest statement
B.S. is a founder of the CellSorter startup company. K.D.P., E.E.J., and E.S.B. are inventors on patent applications regarding the molecules here reported. B.S., K.D.P., E.E.J., and E.S.B. are inventors on a patent application regarding the screening method here developed.
Figures



Similar articles
-
Fluorescent protein engineering by in vivo site-directed mutagenesis.IUBMB Life. 2012 Aug;64(8):684-9. doi: 10.1002/iub.1041. Epub 2012 May 28. IUBMB Life. 2012. PMID: 22639380 Free PMC article.
-
Engineering of Genetically Encoded Bright Near-Infrared Fluorescent Voltage Indicator.Int J Mol Sci. 2025 Feb 8;26(4):1442. doi: 10.3390/ijms26041442. Int J Mol Sci. 2025. PMID: 40003908 Free PMC article.
-
Directed evolution of excited state lifetime and brightness in FusionRed using a microfluidic sorter.Integr Biol (Camb). 2018 Sep 17;10(9):516-526. doi: 10.1039/c8ib00103k. Integr Biol (Camb). 2018. PMID: 30094420 Free PMC article.
-
Ultrahigh-throughput FACS-based screening for directed enzyme evolution.Chembiochem. 2009 Nov 23;10(17):2704-15. doi: 10.1002/cbic.200900384. Chembiochem. 2009. PMID: 19780076 Review.
-
Optogenetic monitoring of membrane potentials.Exp Physiol. 2011 Jan;96(1):13-8. doi: 10.1113/expphysiol.2010.053942. Epub 2010 Sep 17. Exp Physiol. 2011. PMID: 20851856 Review.
Cited by
-
Dual-polarity voltage imaging of the concurrent dynamics of multiple neuron types.Science. 2022 Nov 4;378(6619):eabm8797. doi: 10.1126/science.abm8797. Epub 2022 Nov 4. Science. 2022. PMID: 36378956 Free PMC article.
-
On the fluorescence enhancement of arch neuronal optogenetic reporters.Nat Commun. 2022 Oct 28;13(1):6432. doi: 10.1038/s41467-022-33993-4. Nat Commun. 2022. PMID: 36307417 Free PMC article.
-
Voltage imaging identifies spinal circuits that modulate locomotor adaptation in zebrafish.Neuron. 2022 Apr 6;110(7):1211-1222.e4. doi: 10.1016/j.neuron.2022.01.001. Epub 2022 Jan 31. Neuron. 2022. PMID: 35104451 Free PMC article.
-
Next-generation interfaces for studying neural function.Nat Biotechnol. 2019 Sep;37(9):1013-1023. doi: 10.1038/s41587-019-0198-8. Epub 2019 Aug 12. Nat Biotechnol. 2019. PMID: 31406326 Free PMC article. Review.
-
Cracking the Function of Layers in the Sensory Cortex.Neuron. 2018 Dec 5;100(5):1028-1043. doi: 10.1016/j.neuron.2018.10.032. Neuron. 2018. PMID: 30521778 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

