Unique CD44 intronic SNP is associated with tumor grade in breast cancer: a case control study and in silico analysis
- PMID: 29483847
- PMCID: PMC5824488
- DOI: 10.1186/s12935-018-0522-2
Unique CD44 intronic SNP is associated with tumor grade in breast cancer: a case control study and in silico analysis
Abstract
Background: CD44 encoded by a single gene is a cell surface transmembrane glycoprotein. Exon 2 is one of the important exons to bind CD44 protein to hyaluronan. Experimental evidences show that hyaluronan-CD44 interaction intensifies the proliferation, migration, and invasion of breast cancer cells. Therefore, the current study aimed at investigating the association between specific polymorphisms in exon 2 and its flanking region of CD44 with predisposition to breast cancer.
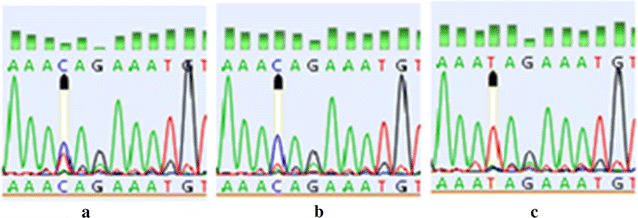
Methods: In the current study, 175 Iranian female patients with breast cancer and 175 age-matched healthy controls were recruited in biobank, Breast Cancer Research Center, Tehran, Iran. Single nucleotide polymorphisms of CD44 exon 2 and its flanking were analyzed via polymerase chain reaction and gene sequencing techniques. Association between the observed variation with breast cancer risk and clinico-pathological characteristics were studied. Subsequently, bioinformatics analysis was conducted to predict potential exonic splicing enhancer (ESE) motifs changed as the result of a mutation.
Results: A unique polymorphism of the gene encoding CD44 was identified at position 14 nucleotide upstream of exon 2 (A37692→G) by the sequencing method. The A > G polymorphism exhibited a significant association with higher-grades of breast cancer, although no significant relation was found between this polymorphism and breast cancer risk. Finally, computational analysis revealed that the intronic mutation generated a new consensus-binding motif for the splicing factor, SC35, within intron 1.
Conclusions: The current study results indicated that A > G polymorphism was associated with breast cancer development; in addition, in silico analysis with ESE finder prediction software showed that the change created a new SC35 binding site.
Keywords: CD44; In silico analysis; Mutation; SR proteins; Single nucleotide polymorphism; Splice variants.
Figures
Similar articles
-
Implications of single nucleotide polymorphisms in CD44 exon 2 for risk of breast cancer.Eur J Cancer Prev. 2011 Sep;20(5):396-402. doi: 10.1097/CEJ.0b013e3283463943. Eur J Cancer Prev. 2011. PMID: 21804359 Free PMC article.
-
CD44 gene polymorphisms in breast cancer risk and prognosis: a study in North Indian population.PLoS One. 2013 Aug 5;8(8):e71073. doi: 10.1371/journal.pone.0071073. Print 2013. PLoS One. 2013. PMID: 23940692 Free PMC article.
-
Multisite and bidirectional exonic splicing enhancer in CD44 alternative exon v3.RNA. 2007 Dec;13(12):2312-23. doi: 10.1261/rna.732807. Epub 2007 Oct 16. RNA. 2007. PMID: 17940137 Free PMC article.
-
CD44: structure, function, and association with the malignant process.Adv Cancer Res. 1997;71:241-319. doi: 10.1016/s0065-230x(08)60101-3. Adv Cancer Res. 1997. PMID: 9111868 Review.
-
Rules and tools to predict the splicing effects of exonic and intronic mutations.Wiley Interdiscip Rev RNA. 2018 Jan;9(1). doi: 10.1002/wrna.1451. Epub 2017 Sep 26. Wiley Interdiscip Rev RNA. 2018. PMID: 28949076 Review.
Cited by
-
Telomere Maintenance Genes are associated with Type 2 Diabetes Susceptibility in Northwest Indian Population Group.Sci Rep. 2020 Apr 15;10(1):6444. doi: 10.1038/s41598-020-63510-w. Sci Rep. 2020. PMID: 32296102 Free PMC article.
-
A SNP involved in alternative splicing of ABCB1 is associated with clopidogrel resistance in coronary heart disease in Chinese population.Aging (Albany NY). 2020 Nov 20;12(24):25684-25699. doi: 10.18632/aging.104177. Epub 2020 Nov 20. Aging (Albany NY). 2020. PMID: 33232268 Free PMC article.
-
Genetic evaluation of the variants using MassARRAY in non-small cell lung cancer among North Indians.Sci Rep. 2021 May 28;11(1):11291. doi: 10.1038/s41598-021-90742-1. Sci Rep. 2021. PMID: 34050209 Free PMC article.
-
Genetic and molecular biology of breast cancer among Iranian patients.J Transl Med. 2019 Jul 8;17(1):218. doi: 10.1186/s12967-019-1968-2. J Transl Med. 2019. PMID: 31286981 Free PMC article. Review.
References
-
- Oskouee MA, Shahmahmoudi S, Nategh R, Esmaeili H-A, Safaeyan F, Moghaddam MZ. Three common TP53 polymorphisms and the risk of breast cancer among groups of Iranian women. Arch Breast Cancer. 2015;2(4):114–119.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous