Inhibition of Mdmx (Mdm4) in vivo induces anti-obesity effects
- PMID: 29484110
- PMCID: PMC5800902
- DOI: 10.18632/oncotarget.23837
Inhibition of Mdmx (Mdm4) in vivo induces anti-obesity effects
Abstract
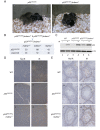
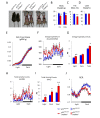
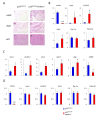
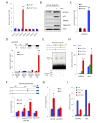
Although cell-cycle arrest, senescence and apoptosis remain as major canonical activities of p53 in tumor suppression, the emerging role of p53 in metabolism has been a topic of great interest. Nevertheless, it is not completely understood how p53-mediated metabolic activities are regulated in vivo and whether this part of the activities has an independent role beyond tumor suppression. Mdmx (also called Mdm4), like Mdm2, acts as a major suppressor of p53 but the embryonic lethality of mdmx-null mice creates difficulties to evaluate its physiological significance in metabolism. Here, we report that the embryonic lethality caused by the deficiency of mdmx, in contrast to the case for mdm2, is fully rescued in the background of p533KR/3KR , an acetylation-defective mutant unable to induce cell-cycle arrest, senescence and apoptosis. p533KR/3KR/mdmx-/- mice are healthy but skinny without obvious developmental defects. p533KR/3KR/mdmx-/- mice are resistant to fat accumulation in adipose tissues upon high fat diet. Notably, the levels of p53 protein are only slightly increased and can be further induced upon DNA damage in p533KR/3KR/mdmx-/- mice, suggesting that Mdmx is only partially required for p53 degradation in vivo. Further analyses indicate that the anti-obesity phenotypes in p533KR/3KR/mdmx-/- mice are caused by activation of lipid oxidation and thermogenic programs in adipose tissues. These results demonstrate the specific effects of the p53/Mdmx axis in lipid metabolism and adipose tissue remodeling and reveal a surprising role of Mdmx inhibition in anti-obesity effects beyond, commonly expected, tumor suppression. Thus, our study has significant implications regarding Mdmx inhibitors in the treatment of obesity related diseases.
Keywords: Mdm4; Mdmx; adipose tissues; metabolism; p53.
Conflict of interest statement
CONFLICTS OF INTEREST The authors declare no competing financial interests.
Figures

References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

