Utility of Glioblastoma Patient-Derived Orthotopic Xenografts in Drug Discovery and Personalized Therapy
- PMID: 29484285
- PMCID: PMC5816058
- DOI: 10.3389/fonc.2018.00023
Utility of Glioblastoma Patient-Derived Orthotopic Xenografts in Drug Discovery and Personalized Therapy
Abstract
Despite substantial effort and resources dedicated to drug discovery and development, new anticancer agents often fail in clinical trials. Among many reasons, the lack of reliable predictive preclinical cancer models is a fundamental one. For decades, immortalized cancer cell cultures have been used to lay the groundwork for cancer biology and the quest for therapeutic responses. However, cell lines do not usually recapitulate cancer heterogeneity or reveal therapeutic resistance cues. With the rapidly evolving exploration of cancer "omics," the scientific community is increasingly investigating whether the employment of short-term patient-derived tumor cell cultures (two- and three-dimensional) and/or patient-derived xenograft models might provide a more representative delineation of the cancer core and its therapeutic response. Patient-derived cancer models allow the integration of genomic with drug sensitivity data on a personalized basis and currently represent the ultimate approach for preclinical drug development and biomarker discovery. The proper use of these patient-derived cancer models might soon influence clinical outcomes and allow the implementation of tailored personalized therapy. When assessing drug efficacy for the treatment of glioblastoma multiforme (GBM), currently, the most reliable models are generated through direct injection of patient-derived cells or more frequently the isolation of glioblastoma cells endowed with stem-like features and orthotopically injecting these cells into the cerebrum of immunodeficient mice. Herein, we present the key strengths, weaknesses, and potential applications of cell- and animal-based models of GBM, highlighting our experience with the glioblastoma stem-like patient cell-derived xenograft model and its utility in drug discovery.
Keywords: glioblastoma; mouse models; patient-derived xenografts; personalized therapy; precision medicine.
Figures
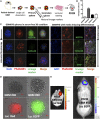
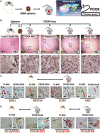
References
-
- Radaelli E, Ceruti R, Patton V, Russo M, Degrassi A, Croci V, et al. Immunohistopathological and neuroimaging characterization of murine orthotopic xenograft models of glioblastoma multiforme recapitulating the most salient features of human disease. Histol Histopathol (2009) 24(7):879–91.10.14670/HH-24.879 - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

