PRRT genomic signature in blood for prediction of 177Lu-octreotate efficacy
- PMID: 29484451
- PMCID: PMC6716527
- DOI: 10.1007/s00259-018-3967-6
PRRT genomic signature in blood for prediction of 177Lu-octreotate efficacy
Abstract
Background: Peptide receptor radionuclide therapy (PRRT) utilizes somatostatin receptor (SSR) overexpression on neuroendocrine tumors (NET) to deliver targeted radiotherapy. Intensity of uptake at imaging is considered related to efficacy but has low sensitivity. A pretreatment strategy to determine individual PRRT response remains a key unmet need. NET transcript expression in blood integrated with tumor grade provides a PRRT predictive quotient (PPQ) which stratifies PRRT "responders" from "non-responders". This study clinically validates the utility of the PPQ in NETs.
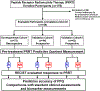
Methods: The development and validation of the PPQ was undertaken in three independent 177Lu-PRRT treated cohorts. Specificity was tested in two separate somatostatin analog-treated cohorts. Prognostic value of the marker was defined in a cohort of untreated patients. The developmental cohort included lung and gastroenteropancreatic [GEP] NETs (n = 72) from IRST Meldola, Italy. The majority were GEP (71%) and low grade (86% G1-G2). Prospective validation cohorts were from Zentralklinik Bad Berka, Germany (n = 44), and Erasmus Medical Center, Rotterdam, Netherlands (n = 42). Each cohort included predominantly well differentiated, low grade (86-95%) lung and GEP-NETs. The non-PRRT comparator cohorts included SSA cohort I, n = 28 (100% low grade, 100% GEP-NET); SSA cohort II, n = 51 (98% low grade; 76% GEP-NET); and an untreated cohort, n = 44 (64% low grade; 91% GEP-NET). Baseline evaluations included clinical information (disease status, grade, SSR) and biomarker (CgA). NET blood gene transcripts (n = 8: growth factor signaling and metabolism) were measured pre-therapy and integrated with tumor Ki67 using a logistic regression model. This provided a binary output: "predicted responder" (PPQ+); "predicted non-responder" (PPQ-). Treatment response was evaluated using RECIST criteria [Responder (stable, partial and complete response) vs Non-Responder)]. Sample measurement and analyses were blinded to study outcome. Statistical evaluation included Kaplan-Meier survival and standard test evaluation analyses.
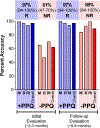
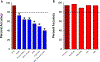
Results: In the developmental cohort, 56% responded to PRRT. The PPQ predicted 100% of responders and 84% of non-responders (accuracy: 93%). In the two validation cohorts (response: 64-79%), the PPQ was 95% accurate (Bad Berka: PPQ + =97%, PPQ- = 93%; Rotterdam: PPQ + =94%, PPQ- = 100%). Overall, the median PFS was not reached in PPQ+ vs PPQ- (10-14 months; HR: 18-77, p < 0.0001). In the comparator cohorts, the predictor (PPQ) was 47-50% accurate for SSA-treatment and 50% as a prognostic. No differences in PFS were respectively noted (PPQ+: 10-12 months vs. PPQ-: 9-15 months).
Conclusion: The PPQ derived from circulating NET specific genes and tumor grade prior to the initiation of therapy is a highly specific predictor of the efficacy of PRRT with an accuracy of 95%.
Keywords: Biomarker; Carcinoid; Liquid biopsy; Neuroendocrine; PRRT; Prediction.
Conflict of interest statement
Figures





References
-
- Modlin IM, Oberg K, Chung DC, Jensen RT, de Herder WW, Thakker RV, et al. Gastroenteropancreatic neuroendocrine tumours. Lancet Oncol. 2008;9:61–72. - PubMed
-
- Pavel M, Valle JW, Eriksson B, Rinke A, Caplin M, Chen J, et al. ENETS consensus guidelines for the standards of care in neuroendocrine neoplasms: systemic therapy - biotherapy and novel targeted agents. Neuroendocrinology. 2017;105:266–80. - PubMed
-
- Faggiano A, Lo Calzo F, Pizza G, Modica R, Colao A. The safety of available treatments options for neuroendocrine tumors. Expert Opin Drug Saf. 2017;16:1149–61. - PubMed
-
- Bodei L, Kwekkeboom DJ, Kidd M, Modlin IM, Krenning EP. Radiolabeled Somatostatin analogue therapy of gastroenteropancreatic cancer. Semin Nucl Med. 2016;46:225–38. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

