Mass spectrometric evidence for neuropeptide-amidating enzymes in Caenorhabditis elegans
- PMID: 29487130
- PMCID: PMC5912480
- DOI: 10.1074/jbc.RA117.000731
Mass spectrometric evidence for neuropeptide-amidating enzymes in Caenorhabditis elegans
Abstract
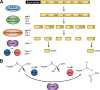
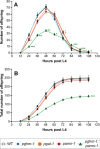
Neuropeptides constitute a vast and functionally diverse family of neurochemical signaling molecules and are widely involved in the regulation of various physiological processes. The nematode Caenorhabditis elegans is well-suited for the study of neuropeptide biochemistry and function, as neuropeptide biosynthesis enzymes are not essential for C. elegans viability. This permits the study of neuropeptide biosynthesis in mutants lacking certain neuropeptide-processing enzymes. Mass spectrometry has been used to study the effects of proprotein convertase and carboxypeptidase mutations on proteolytic processing of neuropeptide precursors and on the peptidome in C. elegans However, the enzymes required for the last step in the production of many bioactive peptides, the carboxyl-terminal amidation reaction, have not been characterized in this manner. Here, we describe three genes that encode homologs of neuropeptide amidation enzymes in C. elegans and used tandem LC-MS to compare neuropeptides in WT animals with those in newly generated mutants for these putative amidation enzymes. We report that mutants lacking both a functional peptidylglycine α-hydroxylating monooxygenase and a peptidylglycine α-amidating monooxygenase had a severely altered neuropeptide profile and also a decreased number of offspring. Interestingly, single mutants of the amidation enzymes still expressed some fully processed amidated neuropeptides, indicating the existence of a redundant amidation mechanism in C. elegans All MS data are available via ProteomeXchange with the identifier PXD008942. In summary, the key steps in neuropeptide processing in C. elegans seem to be executed by redundant enzymes, and loss of these enzymes severely affects brood size, supporting the need of amidated peptides for C. elegans reproduction.
Keywords: Caenorhabditis elegans (C. elegans); PAL; PAM; PHM; amidation; copper monooxygenase; mass spectrometry (MS); neuropeptide; peptides; peptidomics.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures





References
-
- Husson S. J., Reumer A., Temmerman L., De Haes W., Schoofs L., Mertens I., and Baggerman G. (2014) Worm peptidomics. EuPA Open Proteomics 3, 280–290 10.1016/j.euprot.2014.04.005 - DOI
-
- Husson S. J., Clynen E., Baggerman G., Janssen T., and Schoofs L. (2006) Defective processing of neuropeptide precursors in Caenorhabditis elegans lacking proprotein convertase 2 (KPC-2/EGL-3): mutant analysis by mass spectrometry. J. Neurochem. 98, 1999–2012 10.1111/j.1471-4159.2006.04014.x - DOI - PubMed
-
- Husson S. J., Janssen T., Baggerman G., Bogert B., Kahn-Kirby A. H., Ashrafi K., and Schoofs L. (2007) Impaired processing of FLP and NLP peptides in carboxypeptidase E (EGL-21)-deficient Caenorhabditis elegans as analyzed by mass spectrometry. J. Neurochem. 102, 246–260 10.1111/j.1471-4159.2007.04474.x - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

