RNA Interference (RNAi) Screening in Drosophila
- PMID: 29487145
- PMCID: PMC5844339
- DOI: 10.1534/genetics.117.300077
RNA Interference (RNAi) Screening in Drosophila
Abstract
In the last decade, RNA interference (RNAi), a cellular mechanism that uses RNA-guided degradation of messenger RNA transcripts, has had an important impact on identifying and characterizing gene function. First discovered in Caenorhabditis elegans, RNAi can be used to silence the expression of genes through introduction of exogenous double-stranded RNA into cells. In Drosophila, RNAi has been applied in cultured cells or in vivo to perturb the function of single genes or to systematically probe gene function on a genome-wide scale. In this review, we will describe the use of RNAi to study gene function in Drosophila with a particular focus on high-throughput screening methods applied in cultured cells. We will discuss available reagent libraries and cell lines, methodological approaches for cell-based assays, and computational methods for the analysis of high-throughput screens. Furthermore, we will review the generation and use of genome-scale RNAi libraries for tissue-specific knockdown analysis in vivo and discuss the differences and similarities with the use of genome-engineering methods such as CRISPR/Cas9 for functional analysis.
Keywords: Drosophila; FlyBook; RNAi; bioinformatics; functional genomics; genome engineering; high-throughput screening; image-based screening.
Copyright © 2018 Heigwer et al.
Figures

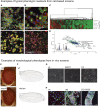

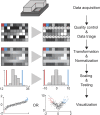
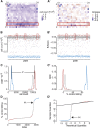


References
-
- Ameres S. L., Zamore P. D., 2013. Diversifying microRNA sequence and function. Nat. Rev. Mol. Cell Biol. 14: 475–488. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

