Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II
- PMID: 29487211
- PMCID: PMC5856558
- DOI: 10.1073/pnas.1722050115
Structural basis of transcriptional stalling and bypass of abasic DNA lesion by RNA polymerase II
Abstract
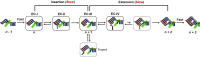
Abasic sites are among the most abundant DNA lesions and interfere with DNA replication and transcription, but the mechanism of their action on transcription remains unknown. Here we applied a combined structural and biochemical approach for a comprehensive investigation of how RNA polymerase II (Pol II) processes an abasic site, leading to slow bypass of lesion. Encounter of Pol II with an abasic site involves two consecutive slow steps: insertion of adenine opposite a noninstructive abasic site (the A-rule), followed by extension of the 3'-rAMP with the next cognate nucleotide. Further studies provided structural insights into the A-rule: ATP is slowly incorporated into RNA in the absence of template guidance. Our structure revealed that ATP is bound to the Pol II active site, whereas the abasic site is located at an intermediate state above the Bridge Helix, a conserved structural motif that is cirtical for Pol II activity. The next extension step occurs in a template-dependent manner where a cognate substrate is incorporated, despite at a much slower rate compared with nondamaged template. During the extension step, neither the cognate substrate nor the template base is located at the canonical position, providing a structural explanation as to why this step is as slow as the insertion step. Taken together, our studies provide a comprehensive understanding of Pol II stalling and bypass of the abasic site in the DNA template.
Keywords: RNA polymerase II; abasic sites in DNA; transcription pausing; transcriptional mutagenesis; translesion transcription.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




References
-
- Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993;362:709–715. - PubMed
-
- Nakamura J, et al. Highly sensitive apurinic/apyrimidinic site assay can detect spontaneous and chemically induced depurination under physiological conditions. Cancer Res. 1998;58:222–225. - PubMed
-
- Loeb LA, Preston BD. Mutagenesis by apurinic/apyrimidinic sites. Annu Rev Genet. 1986;20:201–230. - PubMed
-
- Goodman MF. Error-prone repair DNA polymerases in prokaryotes and eukaryotes. Annu Rev Biochem. 2002;71:17–50. - PubMed
-
- Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu Rev Biochem. 2002;71:133–163. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

