Phosphorylation-dependent activation of the cell wall synthase PBP2a in Streptococcus pneumoniae by MacP
- PMID: 29487215
- PMCID: PMC5856526
- DOI: 10.1073/pnas.1715218115
Phosphorylation-dependent activation of the cell wall synthase PBP2a in Streptococcus pneumoniae by MacP
Abstract
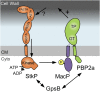
Most bacterial cells are surrounded by an essential cell wall composed of the net-like heteropolymer peptidoglycan (PG). Growth and division of bacteria are intimately linked to the expansion of the PG meshwork and the construction of a cell wall septum that separates the nascent daughter cells. Class A penicillin-binding proteins (aPBPs) are a major family of PG synthases that build the wall matrix. Given their central role in cell wall assembly and importance as drug targets, surprisingly little is known about how the activity of aPBPs is controlled to properly coordinate cell growth and division. Here, we report the identification of MacP (SPD_0876) as a membrane-anchored cofactor of PBP2a, an aPBP synthase of the Gram-positive pathogen Streptococcus pneumoniae We show that MacP localizes to the division site of S. pneumoniae, forms a complex with PBP2a, and is required for the in vivo activity of the synthase. Importantly, MacP was also found to be a substrate for the kinase StkP, a global cell cycle regulator. Although StkP has been implicated in controlling the balance between the elongation and septation modes of cell wall synthesis, none of its substrates are known to modulate PG synthetic activity. Here we show that a phosphoablative substitution in MacP that blocks StkP-mediated phosphorylation prevents PBP2a activity without affecting the MacP-PBP2a interaction. Our results thus reveal a direct connection between PG synthase function and the control of cell morphogenesis by the StkP regulatory network.
Keywords: PBP; bacterial physiology; cell division; cell wall; peptidoglycan.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
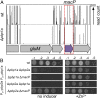
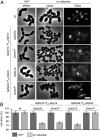
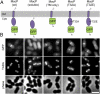

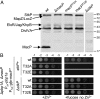
References
-
- Massidda O, Nováková L, Vollmer W. From models to pathogens: How much have we learned about Streptococcus pneumoniae cell division? Environ Microbiol. 2013;15:3133–3157. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

