A validation and extended description of the Lund taxonomy for urothelial carcinoma using the TCGA cohort
- PMID: 29487377
- PMCID: PMC5829240
- DOI: 10.1038/s41598-018-22126-x
A validation and extended description of the Lund taxonomy for urothelial carcinoma using the TCGA cohort
Abstract
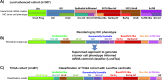
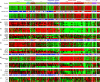
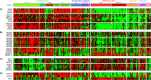
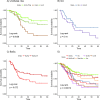
Global gene expression analysis has been a major tool for urothelial carcinoma subtype discovery. This approach has revealed extensive complexity both in intrinsic features of the tumor cells and in the microenvironment. However, global gene expression cannot distinguish between gene expression signals originating from the tumor cells proper and from normal cells in the biopsy. Here, we use a large cohort of advanced urothelial carcinomas for which both gene expression data and extensive immunohistochemistry are available to create a supervised mRNA expression centroid classifier. This classifier identifies the major Lund taxonomy tumor cell phenotypes as defined by IHC. We apply this classifier to the independent TCGA dataset and show excellent associations between identified subtypes and genomic features. We validate a progressed version of Urothelial-like A (UroA-Prog) that shows FGFR3 mutations and CDKN2A deletions, and we show that the variant Urothelial-like C is almost devoid of FGFR3 mutations. We show that Genomically Unstable tumors are very distinct from Urothelial-like tumors at the genomic level, and that tumors classified as Basal/SCC-like all complied with the established definition for Basal/SCC-like tumors. We identify the Mesenchymal-like and Small-cell/Neuroendocrine-like subtypes, and demonstrate that patients with UroB and Sc/NE-like tumors show the worst overall survival.
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
The Lund Molecular Taxonomy Applied to Non-Muscle-Invasive Urothelial Carcinoma.J Mol Diagn. 2022 Sep;24(9):992-1008. doi: 10.1016/j.jmoldx.2022.05.006. Epub 2022 Jul 16. J Mol Diagn. 2022. PMID: 35853574
-
Molecular subtype classification of urothelial carcinoma in Lynch syndrome.Mol Oncol. 2018 Aug;12(8):1286-1295. doi: 10.1002/1878-0261.12325. Epub 2018 Jun 19. Mol Oncol. 2018. PMID: 29791078 Free PMC article.
-
Toward a molecular pathologic classification of urothelial carcinoma.Am J Pathol. 2013 Sep;183(3):681-91. doi: 10.1016/j.ajpath.2013.05.013. Epub 2013 Jul 1. Am J Pathol. 2013. PMID: 23827819
-
[Molecular changes in development and progression of urothelial carcinoma].Verh Dtsch Ges Pathol. 2003;87:172-84. Verh Dtsch Ges Pathol. 2003. PMID: 16888910 Review. German.
-
[Current applications and translational prospects of omics technologies in urothelial carcinoma].Zhonghua Wai Ke Za Zhi. 2024 Nov 1;62(11):1058-1063. doi: 10.3760/cma.j.cn112139-20240814-00375. Zhonghua Wai Ke Za Zhi. 2024. PMID: 39394630 Review. Chinese.
Cited by
-
Tumor Subtyping: Making Sense of Heterogeneity with a Goal Toward Treatment.Bladder Cancer. 2021 Mar 19;7(1):1-11. doi: 10.3233/BLC-200306. eCollection 2021. Bladder Cancer. 2021. PMID: 38993221 Free PMC article. Review.
-
An integrated multi-omics analysis identifies prognostic molecular subtypes of non-muscle-invasive bladder cancer.Nat Commun. 2021 Apr 16;12(1):2301. doi: 10.1038/s41467-021-22465-w. Nat Commun. 2021. PMID: 33863885 Free PMC article.
-
Interruption of the long non-coding RNA HOTAIR signaling axis ameliorates chemotherapy-induced cachexia in bladder cancer.J Biomed Sci. 2022 Dec 6;29(1):104. doi: 10.1186/s12929-022-00887-y. J Biomed Sci. 2022. PMID: 36471329 Free PMC article.
-
A review of the biology and therapeutic implications of cancer-associated fibroblasts (CAFs) in muscle-invasive bladder cancer.Front Oncol. 2022 Oct 13;12:1000888. doi: 10.3389/fonc.2022.1000888. eCollection 2022. Front Oncol. 2022. PMID: 36313650 Free PMC article. Review.
-
Emerging biomarkers in urothelial carcinoma: Challenges and opportunities.Cancer Treat Res Commun. 2020;25:100179. doi: 10.1016/j.ctarc.2020.100179. Epub 2020 May 16. Cancer Treat Res Commun. 2020. PMID: 32920502 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

