Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity
- PMID: 29487387
- PMCID: PMC6127082
- DOI: 10.1038/s41375-018-0067-3
Drugging the lncRNA MALAT1 via LNA gapmeR ASO inhibits gene expression of proteasome subunits and triggers anti-multiple myeloma activity
Abstract
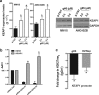
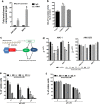
The biological role and therapeutic potential of long non-coding RNAs (lncRNAs) in multiple myeloma (MM) are still to be investigated. Here, we studied the functional significance and the druggability of the oncogenic lncRNA MALAT1 in MM. Targeting MALAT1 by novel LNA-gapmeR antisense oligonucleotide antagonized MM cell proliferation and triggered apoptosis both in vitro and in vivo in a murine xenograft model of human MM. Of note, antagonism of MALAT1 downmodulated the two major transcriptional activators of proteasome subunit genes, namely NRF1 and NRF2, and resulted in reduced trypsin, chymotrypsin and caspase-like proteasome activities and in accumulation of polyubiquitinated proteins. NRF1 and NRF2 decrease upon MALAT1 targeting was due to transcriptional activation of their negative regulator KEAP1, and resulted in reduced expression of anti-oxidant genes and increased ROS levels. In turn, NRF1 promoted MALAT1 expression thus establishing a positive feedback loop. Our findings demonstrate a crucial role of MALAT1 in the regulation of the proteasome machinery, and provide proof-of-concept that its targeting is a novel powerful option for the treatment of MM.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

