The Druggable Pocketome of Corynebacterium diphtheriae: A New Approach for in silico Putative Druggable Targets
- PMID: 29487617
- PMCID: PMC5816920
- DOI: 10.3389/fgene.2018.00044
The Druggable Pocketome of Corynebacterium diphtheriae: A New Approach for in silico Putative Druggable Targets
Abstract
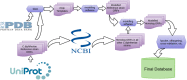
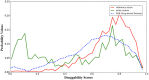
Diphtheria is an acute and highly infectious disease, previously regarded as endemic in nature but vaccine-preventable, is caused by Corynebacterium diphtheriae (Cd). In this work, we used an in silico approach along the 13 complete genome sequences of C. diphtheriae followed by a computational assessment of structural information of the binding sites to characterize the "pocketome druggability." To this end, we first computed the "modelome" (3D structures of a complete genome) of a randomly selected reference strain Cd NCTC13129; that had 13,763 open reading frames (ORFs) and resulted in 1,253 (∼9%) structure models. The amino acid sequences of these modeled structures were compared with the remaining 12 genomes and consequently, 438 conserved protein sequences were obtained. The RCSB-PDB database was consulted to check the template structures for these conserved proteins and as a result, 401 adequate 3D models were obtained. We subsequently predicted the protein pockets for the obtained set of models and kept only the conserved pockets that had highly druggable (HD) values (137 across all strains). Later, an off-target host homology analyses was performed considering the human proteome using NCBI database. Furthermore, the gene essentiality analysis was carried out that gave a final set of 10-conserved targets possessing highly druggable protein pockets. To check the target identification robustness of the pipeline used in this work, we crosschecked the final target list with another in-house target identification approach for C. diphtheriae thereby obtaining three common targets, these were; hisE-phosphoribosyl-ATP pyrophosphatase, glpX-fructose 1,6-bisphosphatase II, and rpsH-30S ribosomal protein S8. Our predicted results suggest that the in silico approach used could potentially aid in experimental polypharmacological target determination in C. diphtheriae and other pathogens, thereby, might complement the existing and new drug-discovery pipelines.
Keywords: Corynebacterium diphtheria; druggable genome; global druggable (GD); highly druggable (HD); pocketome; putative therapeutic targets; structural proteomics.
Figures
Similar articles
-
An integrated structural proteomics approach along the druggable genome of Corynebacterium pseudotuberculosis species for putative druggable targets.BMC Genomics. 2015;16 Suppl 5(Suppl 5):S9. doi: 10.1186/1471-2164-16-S5-S9. Epub 2015 May 26. BMC Genomics. 2015. PMID: 26041381 Free PMC article.
-
An integrative in-silico approach for therapeutic target identification in the human pathogen Corynebacterium diphtheriae.PLoS One. 2017 Oct 19;12(10):e0186401. doi: 10.1371/journal.pone.0186401. eCollection 2017. PLoS One. 2017. PMID: 29049350 Free PMC article.
-
Analysis of the Amino Acid Sequence Variation of the 67-72p Protein and the Structural Pili Proteins of Corynebacterium diphtheriae for their Suitability as Potential Vaccine Antigens.Pol J Microbiol. 2019;68(2):233-246. doi: 10.33073/pjm-2019-025. Pol J Microbiol. 2019. PMID: 31250594 Free PMC article.
-
Corynebacterium diphtheriae: genome diversity, population structure and genotyping perspectives.Infect Genet Evol. 2009 Jan;9(1):1-15. doi: 10.1016/j.meegid.2008.09.011. Epub 2008 Oct 19. Infect Genet Evol. 2009. PMID: 19007916 Review.
-
Evolution, epidemiology and diversity of Corynebacterium diphtheriae: New perspectives on an old foe.Infect Genet Evol. 2016 Sep;43:364-70. doi: 10.1016/j.meegid.2016.06.024. Epub 2016 Jun 9. Infect Genet Evol. 2016. PMID: 27291708 Review.
Cited by
-
An Integrated Database of Small RNAs and Their Interplay With Transcriptional Gene Regulatory Networks in Corynebacteria.Front Microbiol. 2021 Jun 17;12:656435. doi: 10.3389/fmicb.2021.656435. eCollection 2021. Front Microbiol. 2021. PMID: 34220744 Free PMC article.
-
Exploring Nocardia's ecological spectrum and novel therapeutic frontiers through whole-genome sequencing: unraveling drug resistance and virulence factors.Arch Microbiol. 2024 Jan 24;206(2):76. doi: 10.1007/s00203-023-03799-z. Arch Microbiol. 2024. PMID: 38267747
-
Reverse vaccinology and subtractive genomics approaches for identifying common therapeutics against Mycobacterium leprae and Mycobacterium lepromatosis.J Venom Anim Toxins Incl Trop Dis. 2021 Apr 9;27:e20200027. doi: 10.1590/1678-9199-JVATITD-2020-0027. J Venom Anim Toxins Incl Trop Dis. 2021. PMID: 33889182 Free PMC article.
-
Proteomics of Toxigenic Corynebacteria.Proteomes. 2023 Dec 30;12(1):2. doi: 10.3390/proteomes12010002. Proteomes. 2023. PMID: 38250813 Free PMC article. Review.
-
Druggable proteins of Alistipes reveal promising antimicrobial targets against chronic intestinal inflammation.Front Pharmacol. 2025 Apr 24;16:1555062. doi: 10.3389/fphar.2025.1555062. eCollection 2025. Front Pharmacol. 2025. PMID: 40342998 Free PMC article.
References
-
- Barh D., Gupta K., Jain N., Khatri G., Leon-Sicairos N., Canizalez-Roman A., et al. (2013). Conserved host-pathogen PPIs. Globally conserved inter-species bacterial PPIs based conserved host-pathogen interactome derived novel target in C. pseudotuberculosis, C. diphtheriae, M. tuberculosis, C. ulcerans, Y. pestis, and E. coli targeted by Piper betel compounds. Integr. Biol. 5 495–509. 10.1039/c2ib20206a - DOI - PubMed
-
- Barh D., Jain N., Tiwari S., Parida B. P., D’Afonseca V., Li L., et al. (2011). A novel comparative genomics analysis for common drug and vaccine targets in Corynebacterium pseudotuberculosis and other CMN group of human pathogens. Chem. Biol. Drug Des. 78 73–84. 10.1111/j.1747-0285.2011.01118.x - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

