Recognition of DHN-melanin by a C-type lectin receptor is required for immunity to Aspergillus
- PMID: 29489751
- PMCID: PMC5857201
- DOI: 10.1038/nature25974
Recognition of DHN-melanin by a C-type lectin receptor is required for immunity to Aspergillus
Abstract
Resistance to infection is critically dependent on the ability of pattern recognition receptors to recognize microbial invasion and induce protective immune responses. One such family of receptors are the C-type lectins, which are central to antifungal immunity. These receptors activate key effector mechanisms upon recognition of conserved fungal cell-wall carbohydrates. However, several other immunologically active fungal ligands have been described; these include melanin, for which the mechanism of recognition is hitherto undefined. Here we identify a C-type lectin receptor, melanin-sensing C-type lectin receptor (MelLec), that has an essential role in antifungal immunity through recognition of the naphthalene-diol unit of 1,8-dihydroxynaphthalene (DHN)-melanin. MelLec recognizes melanin in conidial spores of Aspergillus fumigatus as well as in other DHN-melanized fungi. MelLec is ubiquitously expressed by CD31+ endothelial cells in mice, and is also expressed by a sub-population of these cells that co-express epithelial cell adhesion molecule and are detected only in the lung and the liver. In mouse models, MelLec was required for protection against disseminated infection with A. fumigatus. In humans, MelLec is also expressed by myeloid cells, and we identified a single nucleotide polymorphism of this receptor that negatively affected myeloid inflammatory responses and significantly increased the susceptibility of stem-cell transplant recipients to disseminated Aspergillus infections. MelLec therefore recognizes an immunologically active component commonly found on fungi and has an essential role in protective antifungal immunity in both mice and humans.
Conflict of interest statement
The authors declare no competing financial interests.
Figures
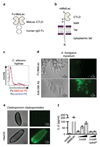
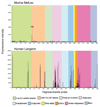

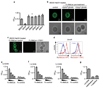
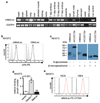
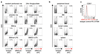
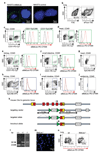
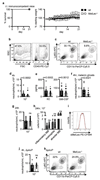
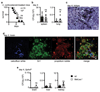
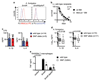
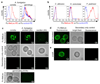
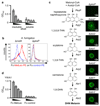
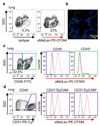
Comment in
-
Melanin triggers antifungal defences.Nature. 2018 Mar 15;555(7696):319-320. doi: 10.1038/d41586-018-02370-x. Nature. 2018. PMID: 29542711 No abstract available.
References
-
- Nosanchuk JD, Casadevall A. The contribution of melanin to microbial pathogenesis. Cell Microbiol. 2003;5:203–223. - PubMed
-
- Pyz E, Brown GD. Screening for ligands of C-type lectin-like receptors. Methods Mol Biol. 2011;748:1–19. - PubMed
-
- Colonna M, Samaridis J, Angman L. Molecular characterization of two novel C-type lectin-like receptors, one of which is selectively expressed in human dendritic cells. Eur J Immunol. 2000;30:697–704. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

