Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor
- PMID: 29489752
- PMCID: PMC6020060
- DOI: 10.1038/nature25786
Regeneration of the lung alveolus by an evolutionarily conserved epithelial progenitor
Abstract
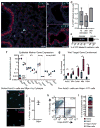
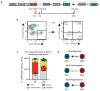
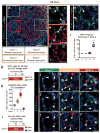
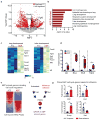
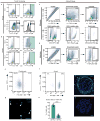
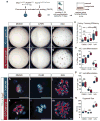
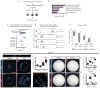
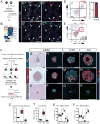
Functional tissue regeneration is required for the restoration of normal organ homeostasis after severe injury. Some organs, such as the intestine, harbour active stem cells throughout homeostasis and regeneration; more quiescent organs, such as the lung, often contain facultative progenitor cells that are recruited after injury to participate in regeneration. Here we show that a Wnt-responsive alveolar epithelial progenitor (AEP) lineage within the alveolar type 2 cell population acts as a major facultative progenitor cell in the distal lung. AEPs are a stable lineage during alveolar homeostasis but expand rapidly to regenerate a large proportion of the alveolar epithelium after acute lung injury. AEPs exhibit a distinct transcriptome, epigenome and functional phenotype and respond specifically to Wnt and Fgf signalling. In contrast to other proposed lung progenitor cells, human AEPs can be directly isolated by expression of the conserved cell surface marker TM4SF1, and act as functional human alveolar epithelial progenitor cells in 3D organoids. Our results identify the AEP lineage as an evolutionarily conserved alveolar progenitor that represents a new target for human lung regeneration strategies.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





Comment in
-
AMP Kinase Activation Attenuates Cardiac Remodeling in Pulmonary Hypertension due to Heart Failure with Preserved Ejection Fraction; Lung Epithelial Progenitor Cells in Alveolar Regeneration; and Drug Discovery and Novel Therapies for Lung Cancer.Am J Respir Cell Mol Biol. 2019 Feb;60(2):244-247. doi: 10.1165/rcmb.2018-0280RO. Am J Respir Cell Mol Biol. 2019. PMID: 30476436 Free PMC article. No abstract available.
-
New insights and therapeutic targets: Lung injury and disease.J Thorac Cardiovasc Surg. 2019 Jan;157(1):416-420. doi: 10.1016/j.jtcvs.2018.09.026. Epub 2018 Oct 6. J Thorac Cardiovasc Surg. 2019. PMID: 30557958 No abstract available.
References
-
- Töpfer L, et al. Influenza A (H1N1) vs non-H1N1 ARDS: analysis of clinical course. J Crit Care. 2014;29:340–346. papers3://publication/doi/10.1016/j.jcrc.2013.12.013. - DOI - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- R03 HL135227/HL/NHLBI NIH HHS/United States
- R01 HL087825/HL/NHLBI NIH HHS/United States
- T32 HL007586/HL/NHLBI NIH HHS/United States
- R01 HL132349/HL/NHLBI NIH HHS/United States
- T32 HL007915/HL/NHLBI NIH HHS/United States
- U01 HL134745/HL/NHLBI NIH HHS/United States
- K23 HL116656/HL/NHLBI NIH HHS/United States
- U01 HL110942/HL/NHLBI NIH HHS/United States
- P30 ES013508/ES/NIEHS NIH HHS/United States
- K12 HD043245/HD/NICHD NIH HHS/United States
- T32 HL007843/HL/NHLBI NIH HHS/United States
- K08 HL140129/HL/NHLBI NIH HHS/United States
- HL129478/NH/NIH HHS/United States
- R01 HL132999/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

