Emerging Technologies for Molecular Diagnosis of Sepsis
- PMID: 29490932
- PMCID: PMC5967692
- DOI: 10.1128/CMR.00089-17
Emerging Technologies for Molecular Diagnosis of Sepsis
Abstract
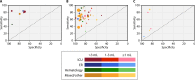
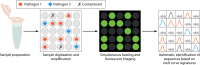
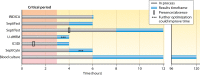
Rapid and accurate profiling of infection-causing pathogens remains a significant challenge in modern health care. Despite advances in molecular diagnostic techniques, blood culture analysis remains the gold standard for diagnosing sepsis. However, this method is too slow and cumbersome to significantly influence the initial management of patients. The swift initiation of precise and targeted antibiotic therapies depends on the ability of a sepsis diagnostic test to capture clinically relevant organisms along with antimicrobial resistance within 1 to 3 h. The administration of appropriate, narrow-spectrum antibiotics demands that such a test be extremely sensitive with a high negative predictive value. In addition, it should utilize small sample volumes and detect polymicrobial infections and contaminants. All of this must be accomplished with a platform that is easily integrated into the clinical workflow. In this review, we outline the limitations of routine blood culture testing and discuss how emerging sepsis technologies are converging on the characteristics of the ideal sepsis diagnostic test. We include seven molecular technologies that have been validated on clinical blood specimens or mock samples using human blood. In addition, we discuss advances in machine learning technologies that use electronic medical record data to provide contextual evaluation support for clinical decision-making.
Keywords: DNA sequencing; biomedical engineering; diagnostic; infectious disease; microbiology techniques.
Copyright © 2018 American Society for Microbiology.
Figures







References
-
- Weinstein MP, Murphy JR, Reller LB, Lichtenstein KA. 1983. The clinical significance of positive blood cultures: a comprehensive analysis of 500 episodes of bacteremia and fungemia in adults. II. Clinical observations, with special reference to factors influencing prognosis. Rev Infect Dis 5:54–70. doi: 10.1093/clinids/5.1.54. - DOI - PubMed
-
- Weinstein MP, Towns ML, Quartey SM, Mirrett S, Reimer LG, Parmigiani G, Reller LB. 1997. The clinical significance of positive blood cultures in the 1990s: a prospective comprehensive evaluation of the microbiology, epidemiology, and outcome of bacteremia and fungemia in adults. Clin Infect Dis 24:584–602. doi: 10.1093/clind/24.4.584. - DOI - PubMed
-
- Elixhauser A, Friedman B, Stranges E. 2011. Septicemia in US hospitals, 2009. Agency for Healthcare Research and Quality, Rockville, MD. - PubMed
-
- Buehler SS, Madison B, Snyder SR, Derzon JH, Cornish NE, Saubolle MA, Weissfeld AS, Weinstein MP, Liebow EB, Wolk DM. 2016. Effectiveness of practices to increase timeliness of providing targeted therapy for inpatients with bloodstream infections: a laboratory medicine best practices systematic review and meta-analysis. Clin Microbiol Rev 29:59–103. doi: 10.1128/CMR.00053-14. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

