Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation
- PMID: 29491135
- PMCID: PMC5859963
- DOI: 10.1101/gad.310367.117
Assessing sufficiency and necessity of enhancer activities for gene expression and the mechanisms of transcription activation
Abstract
Enhancers are important genomic regulatory elements directing cell type-specific transcription. They assume a key role during development and disease, and their identification and functional characterization have long been the focus of scientific interest. The advent of next-generation sequencing and clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9-based genome editing has revolutionized the means by which we study enhancer biology. In this review, we cover recent developments in the prediction of enhancers based on chromatin characteristics and their identification by functional reporter assays and endogenous DNA perturbations. We discuss that the two latter approaches provide different and complementary insights, especially in assessing enhancer sufficiency and necessity for transcription activation. Furthermore, we discuss recent insights into mechanistic aspects of enhancer function, including findings about cofactor requirements and the role of post-translational histone modifications such as monomethylation of histone H3 Lys4 (H3K4me1). Finally, we survey how these approaches advance our understanding of transcription regulation with respect to promoter specificity and transcriptional bursting and provide an outlook covering open questions and promising developments.
Keywords: enhancer prediction; enhancers; gene expression; genetic screens; reporter assays; transcription regulation.
© 2018 Catarino and Stark; Published by Cold Spring Harbor Laboratory Press.
Figures

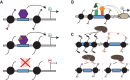

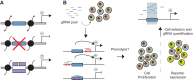
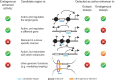
References
-
- Adelman K, Marr MT, Werner J, Saunders A, Ni Z, Andrulis ED, Lis JT. 2005. Efficient release from promoter-proximal stall sites requires transcript cleavage factor TFIIS. Mol Cell 17: 103–112. - PubMed
-
- Akhtar W, de Jong J, Pindyurin AV, Pagie L, Meuleman W, de Ridder J, Berns A, Wessels LFA, van Lohuizen M, van Steensel B. 2013. Chromatin position effects assayed by thousands of reporters integrated in parallel. Cell 154: 914–927. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
