Aspartyl proteases in Candida glabrata are required for suppression of the host innate immune response
- PMID: 29491142
- PMCID: PMC5925793
- DOI: 10.1074/jbc.M117.813741
Aspartyl proteases in Candida glabrata are required for suppression of the host innate immune response
Abstract
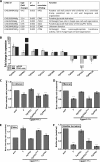
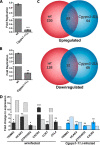
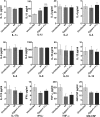
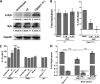
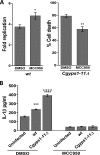
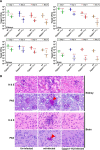
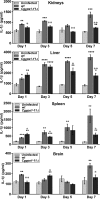
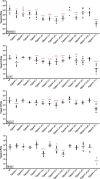
A family of 11 cell surface-associated aspartyl proteases (CgYps1-11), also referred as yapsins, is a key virulence factor in the pathogenic yeast Candida glabrata However, the mechanism by which CgYapsins modulate immune response and facilitate survival in the mammalian host remains to be identified. Here, using RNA-Seq analysis, we report that genes involved in cell wall metabolism are differentially regulated in the Cgyps1-11Δ mutant. Consistently, the mutant contained lower β-glucan and mannan levels and exhibited increased chitin content in the cell wall. As cell wall components are known to regulate the innate immune response, we next determined the macrophage transcriptional response to C. glabrata infection and observed differential expression of genes implicated in inflammation, chemotaxis, ion transport, and the tumor necrosis factor signaling cascade. Importantly, the Cgyps1-11Δ mutant evoked a different immune response, resulting in an enhanced release of the pro-inflammatory cytokine IL-1β in THP-1 macrophages. Further, Cgyps1-11Δ-induced IL-1β production adversely affected intracellular proliferation of co-infected WT cells and depended on activation of spleen tyrosine kinase (Syk) signaling in the host cells. Accordingly, the Syk inhibitor R406 augmented intracellular survival of the Cgyps1-11Δ mutant. Finally, we demonstrate that C. glabrata infection triggers elevated IL-1β production in mouse organs and that the CgYPS genes are required for organ colonization and dissemination in the murine model of systemic infection. Altogether, our results uncover the basis for macrophage-mediated killing of Cgyps1-11Δ cells and provide the first evidence that aspartyl proteases in C. glabrata are required for suppression of IL-1β production in macrophages.
Keywords: IL-1beta; Syk; THP-1 macrophages; Yapsins; cell wall; cell wall remodeling; chemotaxis; interleukin 1 (IL-1); intracellular survival; macrophage; mouse organ colonization; spleen tyrosine kinase (Syk).
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures


Similar articles
-
The yapsin family of aspartyl proteases regulate glucose homeostasis in Candida glabrata.J Biol Chem. 2022 Feb;298(2):101593. doi: 10.1016/j.jbc.2022.101593. Epub 2022 Jan 17. J Biol Chem. 2022. PMID: 35051415 Free PMC article.
-
Global Secretome Characterization of the Pathogenic Yeast Candida glabrata.J Proteome Res. 2020 Jan 3;19(1):49-63. doi: 10.1021/acs.jproteome.9b00299. Epub 2019 Nov 1. J Proteome Res. 2020. PMID: 31621333 Free PMC article.
-
GPI (glycosylphosphatidylinositol)-linked aspartyl proteases regulate vacuole homoeostasis in Candida glabrata.Biochem J. 2014 Mar 1;458(2):323-34. doi: 10.1042/BJ20130757. Biochem J. 2014. PMID: 24341558
-
Intracellular survival of Candida glabrata in macrophages: immune evasion and persistence.FEMS Yeast Res. 2015 Aug;15(5):fov042. doi: 10.1093/femsyr/fov042. Epub 2015 Jun 10. FEMS Yeast Res. 2015. PMID: 26066553 Review.
-
Macrophage pyroptosis induced by Candida albicans.Pathog Dis. 2024 Feb 7;82:ftae003. doi: 10.1093/femspd/ftae003. Pathog Dis. 2024. PMID: 38499444 Free PMC article. Review.
Cited by
-
Using in vivo transcriptomics and RNA enrichment to identify genes involved in virulence of Candida glabrata.Virulence. 2022 Dec;13(1):1285-1303. doi: 10.1080/21505594.2022.2095716. Virulence. 2022. PMID: 35795910 Free PMC article.
-
Aspartyl proteases target host actin nucleator complex protein to limit epithelial innate immunity.EMBO Rep. 2024 Nov;25(11):4846-4875. doi: 10.1038/s44319-024-00270-y. Epub 2024 Sep 30. EMBO Rep. 2024. PMID: 39349750 Free PMC article.
-
Characterization of the Secretome of Pathogenic Candida glabrata and Their Effectiveness against Systemic Candidiasis in BALB/c Mice for Vaccine Development.Pharmaceutics. 2022 Sep 21;14(10):1989. doi: 10.3390/pharmaceutics14101989. Pharmaceutics. 2022. PMID: 36297425 Free PMC article.
-
Candida glabrata Antifungal Resistance and Virulence Factors, a Perfect Pathogenic Combination.Pharmaceutics. 2021 Sep 22;13(10):1529. doi: 10.3390/pharmaceutics13101529. Pharmaceutics. 2021. PMID: 34683822 Free PMC article. Review.
-
Possible Contribution of Alternative Transcript Isoforms in Mature Biofilm Growth Phase of Candida glabrata.Indian J Microbiol. 2022 Dec;62(4):583-601. doi: 10.1007/s12088-022-01036-7. Epub 2022 Aug 27. Indian J Microbiol. 2022. PMID: 36458226 Free PMC article.
References
-
- Brown G. D., Denning D. W., Gow N. A. R., Levitz S. M., Netea M. G., and White T. C. (2012) Hidden killers: human fungal infections. Sci. Transl. Med. 4, 165rv13 - PubMed
-
- Pfaller M. A., Pappas P. G., and Wingard J. R. (2006) Invasive fungal pathogens: current epidemiological trends. Clin. Infect. Dis. 43, S3–S14 10.1086/504490 - DOI
-
- Montagna M. T., Lovero G., Borghi E., Amato G., Andreoni S., Campion L., Lo Cascio G., Lombardi G., Luzzaro F., Manso E., Mussap M., Pecile P., Perin S., Tangorra E., Tronci M., et al. (2014) Candidemia in intensive care unit: a nationwide prospective observational survey (GISIA-3 study) and review of the European literature from 2000 through 2013. Eur. Rev. Med. Pharmacol. Sci. 18, 661–674 - PubMed
-
- Pfaller M. A., Moet G. J., Messer S. A., Jones R. N., and Castanheira M. (2011) Candida bloodstream infections: comparison of species distributions and antifungal resistance patterns in community-onset and nosocomial isolates in the SENTRY Antimicrobial Surveillance Program, 2008–2009. Antimicrob. Agents Chemother. 55, 561–566 10.1128/AAC.01079-10 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

