Relative Abundance of Integrant-Derived Viral RNAs in Infected Tissues Harvested from Chronic Hepatitis B Virus Carriers
- PMID: 29491161
- PMCID: PMC5923063
- DOI: 10.1128/JVI.02221-17
Relative Abundance of Integrant-Derived Viral RNAs in Infected Tissues Harvested from Chronic Hepatitis B Virus Carriers
Abstract
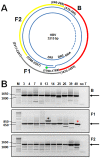
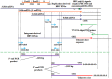
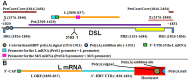
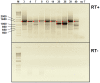
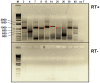
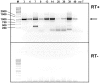
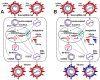
Five matching sets of nonmalignant liver tissues and hepatocellular carcinoma (HCC) samples from individuals chronically infected with hepatitis B virus (HBV) were examined. The HBV genomic sequences were determined by using overlapping PCR amplicons covering the entire viral genome. Four pairs of tissues were infected with HBV genotype C, while one pair was infected with HBV genotype B. HBV replication markers were found in all tissues. In the majority of HCC samples, the levels of pregenomic/precore RNA (pgRNA) and covalently closed circular DNA (cccDNA) were lower than those in liver tissue counterparts. Regardless of the presence of HBV replication markers, (i) integrant-derived HBV RNAs (id-RNAs) were found in all tissues by reverse transcription-PCR (RT-PCR) analysis and were considerably abundant or predominant in 6/10 tissue samples (2 liver and 4 HCC samples), (ii) RNAs that were polyadenylated using the cryptic HBV polyadenylation signal and therefore could be produced by HBV replication or derived from integrated HBV DNA were found in 5/10 samples (3 liver and 2 HCC samples) and were considerably abundant species in 3/10 tissues (2 livers and 1 HCC), and (iii) cccDNA-transcribed RNAs polyadenylated near position 1931 were not abundant in 7/10 tissues (2 liver and 5 HCC samples) and were predominant in only two liver samples. Subsequent RNA sequencing analysis of selected liver/HCC samples also showed relative abundance of id-RNAs in most of the examined tissues. Our findings suggesting that id-RNAs could represent a significant source of HBV envelope proteins, which is independent of viral replication, are discussed in the context of the possible contribution of id-RNAs to the HBV life cycle.IMPORTANCE The relative abundance of integrant-derived HBV RNAs (id-RNAs) in chronically infected tissues suggest that id-RNAs coding for the envelope proteins may facilitate the production of a considerable fraction of surface antigens (HBsAg) in infected cells bearing HBV integrants. If the same cells support HBV replication, then a significant fraction of assembled HBV virions could bear id-RNA-derived HBsAg as a major component of their envelopes. Therefore, the infectivity of these HBV virions and their ability to facilitate virus cell-to-cell spread could be determined mainly by the properties of id-RNA-derived envelope proteins and not by the properties of replication-derived HBsAg. These interpretations suggest that id-RNAs may play a role in the maintenance of chronic HBV infection and therefore contribute to the HBV life cycle. Furthermore, the production of HBsAg from id-RNAs independently of viral replication may explain at least in part why treatment with interferon or nucleos(t)ides in most cases fails to achieve a loss of serum HBsAg.
Keywords: HBV life cycle; chronic HBV infection; hepatitis B virus; integrant-derived viral RNAs.
Copyright © 2018 American Society for Microbiology.
Figures









Similar articles
-
Sera of Individuals Chronically Infected with Hepatitis B Virus (HBV) Contain Diverse RNA Types Produced by HBV Replication or Derived from Integrated HBV DNA.J Virol. 2023 Mar 30;97(3):e0195022. doi: 10.1128/jvi.01950-22. Epub 2023 Mar 6. J Virol. 2023. PMID: 36877036 Free PMC article.
-
Diagnostic Value of Detection of Pregenomic RNA in Sera of Hepatitis B Virus-Infected Patients with Different Clinical Outcomes.J Clin Microbiol. 2020 Jan 28;58(2):e01275-19. doi: 10.1128/JCM.01275-19. Print 2020 Jan 28. J Clin Microbiol. 2020. PMID: 31723011 Free PMC article.
-
Serum hepatitis B surface antigen correlates with tissue covalently closed circular DNA in patients with hepatitis B-associated hepatocellular carcinoma.J Med Virol. 2016 Feb;88(2):244-51. doi: 10.1002/jmv.24326. Epub 2015 Jul 21. J Med Virol. 2016. PMID: 26174662
-
Hepatitis B virus DNA integration: Implications for diagnostics, therapy, and outcome.J Hepatol. 2024 Dec;81(6):1087-1099. doi: 10.1016/j.jhep.2024.06.037. Epub 2024 Jul 5. J Hepatol. 2024. PMID: 38971531 Review.
-
Hepatitis B virus pre-S/S variants in liver diseases.World J Gastroenterol. 2018 Apr 14;24(14):1507-1520. doi: 10.3748/wjg.v24.i14.1507. World J Gastroenterol. 2018. PMID: 29662289 Free PMC article. Review.
Cited by
-
Hepatitis B virus DNA integration as a novel biomarker of hepatitis B virus-mediated pathogenetic properties and a barrier to the current strategies for hepatitis B virus cure.Front Microbiol. 2022 Sep 2;13:972687. doi: 10.3389/fmicb.2022.972687. eCollection 2022. Front Microbiol. 2022. PMID: 36118192 Free PMC article. Review.
-
Sera of Individuals Chronically Infected with Hepatitis B Virus (HBV) Contain Diverse RNA Types Produced by HBV Replication or Derived from Integrated HBV DNA.J Virol. 2023 Mar 30;97(3):e0195022. doi: 10.1128/jvi.01950-22. Epub 2023 Mar 6. J Virol. 2023. PMID: 36877036 Free PMC article.
-
Functional cure of hepatitis B requires silencing covalently closed circular and integrated hepatitis B virus DNA.J Clin Invest. 2022 Sep 15;132(18):e163175. doi: 10.1172/JCI163175. J Clin Invest. 2022. PMID: 36106633 Free PMC article.
-
Enrichment Reveals Extensive Integration of Hepatitis B Virus DNA in Hepatitis Delta Virus-Infected Patients.J Infect Dis. 2024 Sep 23;230(3):e684-e693. doi: 10.1093/infdis/jiae045. J Infect Dis. 2024. PMID: 38271697 Free PMC article.
-
Hepatitis B Virus DNA Integration: In Vitro Models for Investigating Viral Pathogenesis and Persistence.Viruses. 2021 Jan 26;13(2):180. doi: 10.3390/v13020180. Viruses. 2021. PMID: 33530322 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

