Alpha/Beta Interferon (IFN-α/β) Signaling in Astrocytes Mediates Protection against Viral Encephalomyelitis and Regulates IFN-γ-Dependent Responses
- PMID: 29491163
- PMCID: PMC5923078
- DOI: 10.1128/JVI.01901-17
Alpha/Beta Interferon (IFN-α/β) Signaling in Astrocytes Mediates Protection against Viral Encephalomyelitis and Regulates IFN-γ-Dependent Responses
Abstract
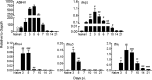
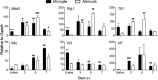
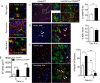
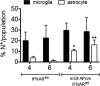
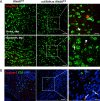
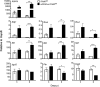
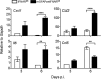
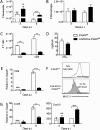
The contribution of distinct central nervous system (CNS) resident cells to protective alpha/beta interferon (IFN-α/β) function following viral infections is poorly understood. Based on numerous immune regulatory functions of astrocytes, we evaluated the contribution of astrocyte IFN-α/β signaling toward protection against the nonlethal glia- and neuronotropic mouse hepatitis virus (MHV) strain A59. Analysis of gene expression associated with IFN-α/β function, e.g., pattern recognition receptors (PRRs) and interferon-stimulated genes (ISGs), revealed lower basal mRNA levels in brain-derived astrocytes than in microglia. Although astrocytes poorly induced Ifnβ mRNA following infection, they upregulated various mRNAs in the IFN-α/β pathway to a higher extent than microglia, supporting effective IFN-α/β responsiveness. Ablation of the IFN-α/β receptor (IFNAR) in astrocytes using mGFAPcre IFNARfl/fl mice resulted in severe encephalomyelitis and mortality, coincident with uncontrolled virus replication. Further, virus spread was not restricted to astrocytes but also affected microglia and neurons, despite increased and sustained Ifnα/β and ISG mRNA levels within the CNS. IFN-γ, a crucial mediator for MHV control, was not impaired in infected mGFAPcre IFNARfl/fl mice despite reduced T cell CNS infiltration. Unexpectedly however, poor induction of IFN-γ-dependent major histocompatibility complex (MHC) class II expression on microglia supported that defective IFN-γ signaling contributes to uncontrolled virus replication. A link between sustained elevated IFN-α/β and impaired responsiveness to IFN-γ supports the novel concept that temporally limited early IFN-α/β responses are critical for effective antiviral IFN-γ function. Overall, our results imply that IFN-α/β signaling in astrocytes is not only critical in limiting early CNS viral spread but also promotes protective antiviral IFN-γ function.IMPORTANCE An antiviral state established by IFN-α/β contains initial viral spread as adaptive immunity develops. While it is apparent that the CNS lacks professional IFN-α/β producers and that resident cells have distinct abilities to elicit innate IFN-α/β responses, protective interactions between inducer and responder cells require further investigation. Infection with a glia- and neuronotropic coronavirus demonstrates that astrocytes mount a delayed but more robust response to infection than microglia, despite their lower basal mRNA levels of IFN-α/β-inducing components. Lethal, uncontrolled viral dissemination following ablation of astrocyte IFN-α/β signaling revealed the importance of IFN-α/β responses in a single cell type for protection. Sustained global IFN-α/β expression associated with uncontrolled virus did not suffice to protect neurons and further impaired responsiveness to protective IFN-γ. The results support astrocytes as critical contributors to innate immunity and the concept that limited IFN-α/β responses are critical for effective subsequent antiviral IFN-γ function.
Keywords: CNS; IFN-α; IFN-β; IFNAR; MHV; astrocytes.
Copyright © 2018 Hwang and Bergmann.
Figures



Similar articles
-
Intercellular Communication Is Key for Protective IFNα/β Signaling During Viral Central Nervous System Infection.Viral Immunol. 2019 Jan/Feb;32(1):1-6. doi: 10.1089/vim.2018.0101. Epub 2018 Sep 15. Viral Immunol. 2019. PMID: 30222502 Free PMC article. Review.
-
Neuronal Ablation of Alpha/Beta Interferon (IFN-α/β) Signaling Exacerbates Central Nervous System Viral Dissemination and Impairs IFN-γ Responsiveness in Microglia/Macrophages.J Virol. 2020 Sep 29;94(20):e00422-20. doi: 10.1128/JVI.00422-20. Print 2020 Sep 29. J Virol. 2020. PMID: 32796063 Free PMC article.
-
Myd88 Initiates Early Innate Immune Responses and Promotes CD4 T Cells during Coronavirus Encephalomyelitis.J Virol. 2015 Sep;89(18):9299-312. doi: 10.1128/JVI.01199-15. Epub 2015 Jul 1. J Virol. 2015. PMID: 26136579 Free PMC article.
-
Ifit2 deficiency results in uncontrolled neurotropic coronavirus replication and enhanced encephalitis via impaired alpha/beta interferon induction in macrophages.J Virol. 2014 Jan;88(2):1051-64. doi: 10.1128/JVI.02272-13. Epub 2013 Nov 6. J Virol. 2014. PMID: 24198415 Free PMC article.
-
Fine Tuning the Cytokine Storm by IFN and IL-10 Following Neurotropic Coronavirus Encephalomyelitis.Front Immunol. 2018 Dec 20;9:3022. doi: 10.3389/fimmu.2018.03022. eCollection 2018. Front Immunol. 2018. PMID: 30619363 Free PMC article. Review.
Cited by
-
Zika Virus Infection Disrupts Astrocytic Proteins Involved in Synapse Control and Axon Guidance.Front Microbiol. 2019 Mar 26;10:596. doi: 10.3389/fmicb.2019.00596. eCollection 2019. Front Microbiol. 2019. PMID: 30984137 Free PMC article.
-
Emerging Roles of Type-I Interferons in Neuroinflammation, Neurological Diseases, and Long-Haul COVID.Int J Mol Sci. 2022 Nov 19;23(22):14394. doi: 10.3390/ijms232214394. Int J Mol Sci. 2022. PMID: 36430870 Free PMC article. Review.
-
Distinct Gene Profiles of Bone Marrow-Derived Macrophages and Microglia During Neurotropic Coronavirus-Induced Demyelination.Front Immunol. 2018 Jun 11;9:1325. doi: 10.3389/fimmu.2018.01325. eCollection 2018. Front Immunol. 2018. PMID: 29942315 Free PMC article.
-
Intercellular Communication Is Key for Protective IFNα/β Signaling During Viral Central Nervous System Infection.Viral Immunol. 2019 Jan/Feb;32(1):1-6. doi: 10.1089/vim.2018.0101. Epub 2018 Sep 15. Viral Immunol. 2019. PMID: 30222502 Free PMC article. Review.
-
Expression of SARS-CoV-2-related receptors in cells of the neurovascular unit: implications for HIV-1 infection.J Neuroinflammation. 2021 Jul 29;18(1):167. doi: 10.1186/s12974-021-02210-2. J Neuroinflammation. 2021. PMID: 34325716 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

