Gut microbial composition in patients with psoriasis
- PMID: 29491401
- PMCID: PMC5830498
- DOI: 10.1038/s41598-018-22125-y
Gut microbial composition in patients with psoriasis
Abstract
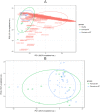
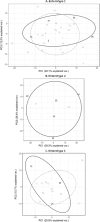
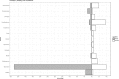
Since the last 5-10 years the relevance of the gut microbiome on different intestinal illnesses has been revealed. Recent findings indicate the effect of gut microbiome on certain dermatological diseases such as atopic dermatitis. However, data on other skin diseases such as psoriasis are limited. This is the first time attempting to reveal the gut microbiome composition of psoriatic patients with a prospective study including a group of patients with plaque psoriasis, analyzing their gut microbiome and the relationship between the microbiome composition and bacterial translocation. The microbiome of a cohort of 52 psoriatic patients (PASI score ≥6) was obtained by 16s rRNA massive sequencing with MiSeq platform (Illumina inc, San Diego) with an average of 85,000 sequences per sample. The study of the gut microbiome and enterotype shows from the first time a specific "psoriatic core intestinal microbiome" that clearly differs from the one present in healthy population. In addition, those psoriatic patients classified as belonging to enterotype 2 tended to experience more frequent bacterial translocation and higher inflammatory status (71%) than patients with other enterotypes (16% for enterotype 1; and 21% for enterotype 3).
Conflict of interest statement
D.R., S.G. and E.C. are employees of Biopolis, S.L.-ADM. F.C., E.C.L. and D.R. are employees of Lifesequencing, S.L.-ADM. M.C.G. is an employee of Especialidades Farmacéuticas Centrum. J.M.P.-O. is an employee of Korott. V.N.L., A.R.B., J.H.P. and M.G. state no conflict of interest.
Figures
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

