Sheep genome functional annotation reveals proximal regulatory elements contributed to the evolution of modern breeds
- PMID: 29491421
- PMCID: PMC5830443
- DOI: 10.1038/s41467-017-02809-1
Sheep genome functional annotation reveals proximal regulatory elements contributed to the evolution of modern breeds
Abstract
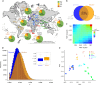
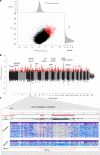
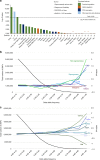
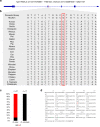
Domestication fundamentally reshaped animal morphology, physiology and behaviour, offering the opportunity to investigate the molecular processes driving evolutionary change. Here we assess sheep domestication and artificial selection by comparing genome sequence from 43 modern breeds (Ovis aries) and their Asian mouflon ancestor (O. orientalis) to identify selection sweeps. Next, we provide a comparative functional annotation of the sheep genome, validated using experimental ChIP-Seq of sheep tissue. Using these annotations, we evaluate the impact of selection and domestication on regulatory sequences and find that sweeps are significantly enriched for protein coding genes, proximal regulatory elements of genes and genome features associated with active transcription. Finally, we find individual sites displaying strong allele frequency divergence are enriched for the same regulatory features. Our data demonstrate that remodelling of gene expression is likely to have been one of the evolutionary forces that drove phenotypic diversification of this common livestock species.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

