Heritable DNA methylation marks associated with susceptibility to breast cancer
- PMID: 29491469
- PMCID: PMC5830448
- DOI: 10.1038/s41467-018-03058-6
Heritable DNA methylation marks associated with susceptibility to breast cancer
Abstract
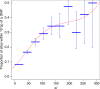
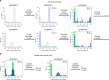
Mendelian-like inheritance of germline DNA methylation in cancer susceptibility genes has been previously reported. We aimed to scan the genome for heritable methylation marks associated with breast cancer susceptibility by studying 25 Australian multiple-case breast cancer families. Here we report genome-wide DNA methylation measured in 210 peripheral blood DNA samples provided by family members using the Infinium HumanMethylation450. We develop and apply a new statistical method to identify heritable methylation marks based on complex segregation analysis. We estimate carrier probabilities for the 1000 most heritable methylation marks based on family structure, and we use Cox proportional hazards survival analysis to identify 24 methylation marks with corresponding carrier probabilities significantly associated with breast cancer. We replicate an association with breast cancer risk for four of the 24 marks using an independent nested case-control study. Here, we report a novel approach for identifying heritable DNA methylation marks associated with breast cancer risk.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

