Elucidating Duramycin's Bacterial Selectivity and Mode of Action on the Bacterial Cell Envelope
- PMID: 29491859
- PMCID: PMC5817074
- DOI: 10.3389/fmicb.2018.00219
Elucidating Duramycin's Bacterial Selectivity and Mode of Action on the Bacterial Cell Envelope
Abstract
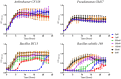
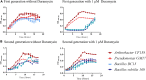
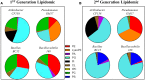
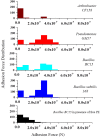
The use of naturally occurring antimicrobial peptides provides a promising route to selectively target pathogenic agents and to shape microbiome structure. Lantibiotics, such as duramycin, are one class of bacterially produced peptidic natural products that can selectively inhibit the growth of other bacteria. However, despite longstanding characterization efforts, the microbial selectivity and mode of action of duramycin are still obscure. We describe here a suite of biological, chemical, and physical characterizations that shed new light on the selective and mechanistic aspects of duramycin activity. Bacterial screening assays have been performed using duramycin and Populus-derived bacterial isolates to determine species selectivity. Lipidomic profiles of selected resistant and sensitive strains show that the sensitivity of Gram-positive bacteria depends on the presence of phosphatidylethanolamine (PE) in the cell membrane. Further the surface and interface morphology were studied by high resolution atomic force microscopy and showed a progression of cellular changes in the cell envelope after treatment with duramycin for the susceptible bacterial strains. Together, these molecular and cellular level analyses provide insight into duramycin's mode of action and a better understanding of its selectivity.
Keywords: atomic force microscopy (AFM); cell elasticity; duramycin; lipid; lipidomics; molecular adhesion force; peptidoglycan; phosphatidylethanolamine (PE).
Figures

References
-
- Arnison P. G., Bibb M. J., Bierbaum G., Bowers A. A., Bugni T. S., Bulaj G., et al. (2013). Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat. Prod. Rep. 30 108–160. 10.1039/c2np20085f - DOI - PMC - PubMed
-
- Baddiley J. (1972). Teichoic acids in cell walls and membranes of bacteria. Essays Biochem. 8 35–77. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

