A Comprehensive Atlas of E3 Ubiquitin Ligase Mutations in Neurological Disorders
- PMID: 29491882
- PMCID: PMC5817383
- DOI: 10.3389/fgene.2018.00029
A Comprehensive Atlas of E3 Ubiquitin Ligase Mutations in Neurological Disorders
Abstract
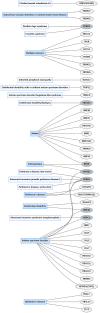
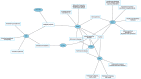
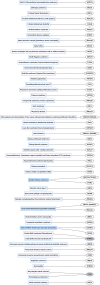
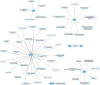
Protein ubiquitination is a posttranslational modification that plays an integral part in mediating diverse cellular functions. The process of protein ubiquitination requires an enzymatic cascade that consists of a ubiquitin activating enzyme (E1), ubiquitin conjugating enzyme (E2) and an E3 ubiquitin ligase (E3). There are an estimated 600-700 E3 ligase genes representing ~5% of the human genome. Not surprisingly, mutations in E3 ligase genes have been observed in multiple neurological conditions. We constructed a comprehensive atlas of disrupted E3 ligase genes in common (CND) and rare neurological diseases (RND). Of the predicted and known human E3 ligase genes, we found ~13% were mutated in a neurological disorder with 83 total genes representing 70 different types of neurological diseases. Of the E3 ligase genes identified, 51 were associated with an RND. Here, we provide an updated list of neurological disorders associated with E3 ligase gene disruption. We further highlight research in these neurological disorders and discuss the advanced technologies used to support these findings.
Keywords: angelman syndrome; neurological; rare diseases; transgenic; ubiquitin.
Figures

References
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

