Efficient iterative virtual screening with Apache Spark and conformal prediction
- PMID: 29492726
- PMCID: PMC5833896
- DOI: 10.1186/s13321-018-0265-z
Efficient iterative virtual screening with Apache Spark and conformal prediction
Abstract
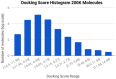
Background: Docking and scoring large libraries of ligands against target proteins forms the basis of structure-based virtual screening. The problem is trivially parallelizable, and calculations are generally carried out on computer clusters or on large workstations in a brute force manner, by docking and scoring all available ligands.
Contribution: In this study we propose a strategy that is based on iteratively docking a set of ligands to form a training set, training a ligand-based model on this set, and predicting the remainder of the ligands to exclude those predicted as 'low-scoring' ligands. Then, another set of ligands are docked, the model is retrained and the process is repeated until a certain model efficiency level is reached. Thereafter, the remaining ligands are docked or excluded based on this model. We use SVM and conformal prediction to deliver valid prediction intervals for ranking the predicted ligands, and Apache Spark to parallelize both the docking and the modeling.
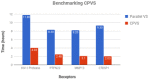
Results: We show on 4 different targets that conformal prediction based virtual screening (CPVS) is able to reduce the number of docked molecules by 62.61% while retaining an accuracy for the top 30 hits of 94% on average and a speedup of 3.7. The implementation is available as open source via GitHub ( https://github.com/laeeq80/spark-cpvs ) and can be run on high-performance computers as well as on cloud resources.
Keywords: Apache Spark; Cloud computing; Conformal prediction; Docking; Virtual screening.
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources

