Potential Roles for G-Quadruplexes in Mitochondria
- PMID: 29493440
- PMCID: PMC6113130
- DOI: 10.2174/0929867325666180228165527
Potential Roles for G-Quadruplexes in Mitochondria
Abstract
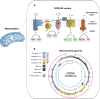
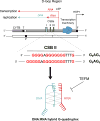
Some DNA or RNA sequences rich in guanine (G) nucleotides can adopt noncanonical conformations known as G-quadruplexes (G4). In the nuclear genome, G4 motifs have been associated with genome instability and gene expression defects, but they are increasingly recognized to be regulatory structures. Recent studies have revealed that G4 structures can form in the mitochondrial genome (mtDNA) and potential G4 forming sequences are associated with the origin of mtDNA deletions. However, little is known about the regulatory role of G4 structures in mitochondria. In this short review, we will explore the potential for G4 structures to regulate mitochondrial function, based on evidence from the nucleus.
Keywords: G-quadruplexes; G4 ligand; mitochondrial gene expression; mitochondrial genome instability; mtDNA; mtDNA deletions.; mtDNA depletion.
Copyright© Bentham Science Publishers; For any queries, please email at epub@benthamscience.net.
Figures



References
-
- Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. - PubMed
-
- Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature 1990 Mar 29; - PubMed
-
- Hazel P, Parkinson GN, Neidle S. Topology variation and loop structural homology in crystal and simulated structures of a bimolecular DNA quadruplex. J. Am. Chem. Soc. 2006;128:5480–7. - PubMed
-
- Karsisiotis AI, Hessari NM, Novellino E, Spada GP, Randazzo A, Webba da Silva M. Topological characterization of nucleic acid G-quadruplexes by UV absorption and circular dichroism. Angew. Chem. Int. Ed. Engl. 2011;50:10645–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
