Translesion and Repair DNA Polymerases: Diverse Structure and Mechanism
- PMID: 29494238
- PMCID: PMC6098713
- DOI: 10.1146/annurev-biochem-062917-012405
Translesion and Repair DNA Polymerases: Diverse Structure and Mechanism
Abstract
The number of DNA polymerases identified in each organism has mushroomed in the past two decades. Most newly found DNA polymerases specialize in translesion synthesis and DNA repair instead of replication. Although intrinsic error rates are higher for translesion and repair polymerases than for replicative polymerases, the specialized polymerases increase genome stability and reduce tumorigenesis. Reflecting the numerous types of DNA lesions and variations of broken DNA ends, translesion and repair polymerases differ in structure, mechanism, and function. Here, we review the unique and general features of polymerases specialized in lesion bypass, as well as in gap-filling and end-joining synthesis.
Keywords: A-family; B-family; DSBs; NHEJ; TLS; TMEJ; X-family; Y-family; proofreading; sGRS; small gap-filling repair synthesis.
Figures



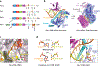
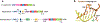
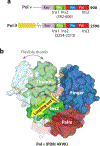

References
-
- Bessman MJ, Kornberg A, Lehman IR, Simms ES. 1956. Enzymic synthesis of deoxyribonucleic acid. Biochim Biophys Acta 21: 197–8 - PubMed
-
- Englund PT, Deutscher MP, Jovin TM, Kelly RB, Cozzarelli NR, Kornberg A. 1968. Structural and functional properties of Escherichia coli DNA polymerase. Cold Spring Harb Symp Quant Biol 33: 1–9 - PubMed
-
- Ollis DL, Brick P, Hamlin R, Xuong NG, Steitz TA. 1985. Structure of large fragment of Escherichia coli DNA polymerase I complexed with dTMP. Nature 313: 762–6 - PubMed
-
- Lindahl T 2001. Keynote: past, present, and future aspects of base excision repair. Prog Nucleic Acid Res Mol Biol 68: xvii–xxx - PubMed
-
- Setlow RB. 1966. Cyclobutane-type pyrimidine dimers in polynucleotides. Science 153: 379–86 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

