Bacillus Cellulase Molecular Cloning, Expression, and Surface Display on the Outer Membrane of Escherichia coli
- PMID: 29495265
- PMCID: PMC6017809
- DOI: 10.3390/molecules23020503
Bacillus Cellulase Molecular Cloning, Expression, and Surface Display on the Outer Membrane of Escherichia coli
Abstract
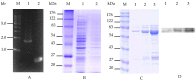
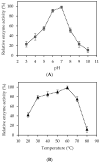
One of the main challenges of using recombinant enzymes is that they are derived from genetically-modified microorganisms commonly located in the intracellular region. The use of these recombinant enzymes for commercial purposes requires the additional processes of cell disruption and purification, which may result in enzyme loss, denaturation, and increased total production cost. In this study, the cellulase gene of Bacillus licheniformis ATCC 14580 was cloned, over-expressed, and surface displayed in recombinant Escherichia coli using an ice-nucleation protein (INP). INP, an outer membrane-bound protein from Pseudomonas syringae, was utilized as an anchor linker, which was cloned with a foreign cellulase gene into the pET21a vector to develop a surface display system on the outer membrane of E. coli. The resulting strain successfully revealed cellulase on the host cell surface. The over-expressed INP-cellulase fusion protein was confirmed via staining assay for determining the extracellular cellulase and Western blotting method for the molecular weight (MW) of cellulase, which was estimated to be around 61.7 kDa. Cell fractionation and localization tests demonstrated that the INP-cellulase fusion protein was mostly present in the supernatant (47.5%) and outer membrane (19.4%), while the wild-type strain intracellularly retained enzymes within cytosol (>61%), indicating that the INP gene directed the cellulase expression on the bacteria cell surface. Further studies of the optimal enzyme activity were observed at 60 °C and pH 7.0, and at least 75% of maximal enzyme activity was preserved at 70 °C.
Keywords: Bacillus licheniformis; Pseudomonas syringae; cellulase; ice nucleation protein; surface anchoring; whole cell catalysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Feng Y., Zhang M., Mujumdar A.S., Gao Z. Recent research process of fermented plant extract: A review. Trends Food Sci. Technol. 2017;65:40–48. doi: 10.1016/j.tifs.2017.04.006. - DOI
-
- Hasunuma T., Okazaki F., Okai N., Hara K.Y., Ishii J., Kondo A. A review of enzymes and microbes for lignocellulosic biorefinery and the possibility of their application to consolidated bioprocessing technology. Bioresour. Technol. 2013;135:513–522. doi: 10.1016/j.biortech.2012.10.047. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

