Plant Mitochondrial Inner Membrane Protein Insertion
- PMID: 29495281
- PMCID: PMC5855863
- DOI: 10.3390/ijms19020641
Plant Mitochondrial Inner Membrane Protein Insertion
Abstract
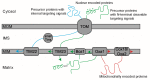
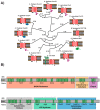
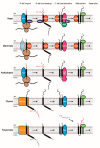
During the biogenesis of the mitochondrial inner membrane, most nuclear-encoded inner membrane proteins are laterally released into the membrane by the TIM23 and the TIM22 machinery during their import into mitochondria. A subset of nuclear-encoded mitochondrial inner membrane proteins and all the mitochondrial-encoded inner membrane proteins use the Oxa machinery-which is evolutionarily conserved from the endosymbiotic bacterial ancestor of mitochondria-for membrane insertion. Compared to the mitochondria from other eukaryotes, plant mitochondria have several unique features, such as a larger genome and a branched electron transport pathway, and are also involved in additional cellular functions such as photorespiration and stress perception. This review focuses on the unique aspects of plant mitochondrial inner membrane protein insertion machinery, which differs from that in yeast and humans, and includes a case study on the biogenesis of Cox2 in yeast, humans, two plant species, and an algal species to highlight lineage-specific similarities and differences. Interestingly, unlike mitochondria of other eukaryotes but similar to bacteria and chloroplasts, plant mitochondria appear to use the Tat machinery for membrane insertion of the Rieske Fe/S protein.
Keywords: Oxa; membrane insertion; plant mitochondria; twin-arginine translocation.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Sloan D.B., Alverson A.J., Chuckalovcak J.P., Wu M., McCauley D.E., Palmer J.D., Taylor D.R. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012;10:e1001241. doi: 10.1371/journal.pbio.1001241. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

