Complete Mitochondrial Genome of Suwallia teleckojensis (Plecoptera: Chloroperlidae) and Implications for the Higher Phylogeny of Stoneflies
- PMID: 29495588
- PMCID: PMC5877541
- DOI: 10.3390/ijms19030680
Complete Mitochondrial Genome of Suwallia teleckojensis (Plecoptera: Chloroperlidae) and Implications for the Higher Phylogeny of Stoneflies
Abstract
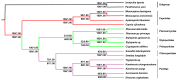
Stoneflies comprise an ancient group of insects, but the phylogenetic position of Plecoptera and phylogenetic relations within Plecoptera have long been controversial, and more molecular data is required to reconstruct precise phylogeny. Herein, we present the complete mitogenome of a stonefly, Suwallia teleckojensis, which is 16146 bp in length and consists of 13 protein-coding genes (PCGs), 2 ribosomal RNAs (rRNAs), 22 transfer RNAs (tRNAs) and a control region (CR). Most PCGs initiate with the standard start codon ATN. However, ND5 and ND1 started with GTG and TTG. Typical termination codons TAA and TAG were found in eleven PCGs, and the remaining two PCGs (COII and ND5) have incomplete termination codons. All transfer RNA genes (tRNAs) have the classic cloverleaf secondary structures, with the exception of tRNASer(AGN), which lacks the dihydrouridine (DHU) arm. Secondary structures of the two ribosomal RNAs were shown referring to previous models. A large tandem repeat region, two potential stem-loop (SL) structures, Poly N structure (2 poly-A, 1 poly-T and 1 poly-C), and four conserved sequence blocks (CSBs) were detected in the control region. Finally, both maximum likelihood (ML) and Bayesian inference (BI) analyses suggested that the Capniidae was monophyletic, and the other five stonefly families form a monophyletic group. In this study, S. teleckojensis was closely related to Sweltsa longistyla, and Chloroperlidae and Perlidae were herein supported to be a sister group.
Keywords: Chloroperlidae; Plecoptera; Suwallia teleckojensis; mitochondrial genome; phylogenetics.
Conflict of interest statement
The authors declare no conflict of interest.
Figures







Similar articles
-
The complete mitochondrial genome of a stonefly species, Suwallia bimaculata (Plecoptera: Chloroperlidae).Mitochondrial DNA B Resour. 2019 Sep 2;4(2):2828-2829. doi: 10.1080/23802359.2019.1660263. Mitochondrial DNA B Resour. 2019. PMID: 33365747 Free PMC article.
-
The first two mitochondrial genomes from Taeniopterygidae (Insecta: Plecoptera): Structural features and phylogenetic implications.Int J Biol Macromol. 2018 May;111:70-76. doi: 10.1016/j.ijbiomac.2017.12.150. Epub 2017 Dec 29. Int J Biol Macromol. 2018. PMID: 29292150
-
Molecular phylogeny inferred from the mitochondrial genomes of Plecoptera with Oyamia nigribasis (Plecoptera: Perlidae).Sci Rep. 2020 Dec 1;10(1):20955. doi: 10.1038/s41598-020-78082-y. Sci Rep. 2020. PMID: 33262442 Free PMC article.
-
The mitochondrial genome of Leuctra sp. (Plecoptera: Leuctridae) and its performance in phylogenetic analyses.Zootaxa. 2019 Sep 19;4671(4):zootaxa.4671.4.8. doi: 10.11646/zootaxa.4671.4.8. Zootaxa. 2019. PMID: 31716036
-
The first mitochondrial genome from Scopuridae (Insecta: Plecoptera) reveals structural features and phylogenetic implications.Int J Biol Macromol. 2019 Feb 1;122:893-902. doi: 10.1016/j.ijbiomac.2018.11.019. Epub 2018 Nov 6. Int J Biol Macromol. 2019. PMID: 30412754
Cited by
-
Two Complete Mitochondrial Genomes From Leuctridae (Plecoptera: Nemouroidea): Implications for the Phylogenetic Relationships Among Stoneflies.J Insect Sci. 2021 Jan 1;21(1):16. doi: 10.1093/jisesa/ieab009. J Insect Sci. 2021. PMID: 33590866 Free PMC article.
-
Tick mitochondrial genomes: structural characteristics and phylogenetic implications.Parasit Vectors. 2019 Sep 13;12(1):451. doi: 10.1186/s13071-019-3705-3. Parasit Vectors. 2019. PMID: 31519208 Free PMC article. Review.
-
Characterization of the Complete Mitochondrial Genome of Harpalus sinicus and Its Implications for Phylogenetic Analyses.Genes (Basel). 2019 Sep 18;10(9):724. doi: 10.3390/genes10090724. Genes (Basel). 2019. PMID: 31540431 Free PMC article.
-
Mitochondrial genomes of the stoneflies Mesonemourametafiligera and Mesonemouratritaenia (Plecoptera, Nemouridae), with a phylogenetic analysis of Nemouroidea.Zookeys. 2019 Apr 4;835:43-63. doi: 10.3897/zookeys.835.32470. eCollection 2019. Zookeys. 2019. PMID: 31043849 Free PMC article.
-
The complete mitochondrial genome of a stonefly species, Suwallia bimaculata (Plecoptera: Chloroperlidae).Mitochondrial DNA B Resour. 2019 Sep 2;4(2):2828-2829. doi: 10.1080/23802359.2019.1660263. Mitochondrial DNA B Resour. 2019. PMID: 33365747 Free PMC article.
References
-
- Wolstenholme D.R. Animal mitochondrial DNA: Structure and evolution. Int. Rev. Cytol. 1992;141:173–216. - PubMed
-
- Fochetti R., Figueroa J.M.T. Global diversity of stoneflies (Plecoptera, Insecta) in freshwater. Hydrobiologia. 2008;595:365–377. doi: 10.1007/s10750-007-9031-3. - DOI
-
- Plecoptera Species File Online. [(accessed on 21 January 2018)]; Available online: http://Plecoptera.SpeciesFile.org.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

