Quantitative Phosphoproteomic and Metabolomic Analyses Reveal GmMYB173 Optimizes Flavonoid Metabolism in Soybean under Salt Stress
- PMID: 29496908
- PMCID: PMC5986248
- DOI: 10.1074/mcp.RA117.000417
Quantitative Phosphoproteomic and Metabolomic Analyses Reveal GmMYB173 Optimizes Flavonoid Metabolism in Soybean under Salt Stress
Abstract
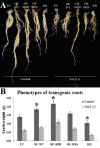
Salinity causes osmotic stress to crops and limits their productivity. To understand the mechanism underlying soybean salt tolerance, proteomics approach was used to identify phosphoproteins altered by NaCl treatment. Results revealed that 412 of the 4698 quantitatively analyzed phosphopeptides were significantly up-regulated on salt treatment, including a phosphopeptide covering the serine 59 in the transcription factor GmMYB173. Our data showed that GmMYB173 is one of the three MYB proteins differentially phosphorylated on salt treatment, and a substrate of the casein kinase-II. MYB recognition sites exist in the promoter of flavonoid synthase gene GmCHS5 and one was found to mediate its recognition by GmMYB173, an event facilitated by phosphorylation. Because GmCHS5 catalyzes the synthesis of chalcone, flavonoids derived from chalcone were monitored using metabolomics approach. Results revealed that 24 flavonoids of 6745 metabolites were significantly up-regulated after salt treatment. We further compared the salt tolerance and flavonoid accumulation in soybean transgenic roots expressing the 35S promoter driven cds and RNAi constructs of GmMYB173 and GmCHS5, as well as phospho-mimic (GmMYB173S59D ) and phospho-ablative (GmMYB173S59A ) mutants of GmMYB173 Overexpression of GmMYB173S59D and GmCHS5 resulted in the highest increase in salt tolerance and accumulation of cyaniding-3-arabinoside chloride, a dihydroxy B-ring flavonoid. The dihydroxy B-ring flavonoids are more effective as anti-oxidative agents when compared with monohydroxy B-ring flavonoids, such as formononetin. Hence the salt-triggered phosphorylation of GmMYB173, subsequent increase in its affinity to GmCHS5 promoter and the elevated transcription of GmCHS5 likely contribute to soybean salt tolerance by enhancing the accumulation of dihydroxy B-ring flavonoids.
Keywords: Flavonoid Metabolism; GmMYB173; Metabolomics; Metabonomics; Oxidative stress; Phosphoproteome; Phosphoproteomics; Protein Modification*; Soybean; Stress response; iTRAQ.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Figures






References
-
- Sobhanian H., Aghaei K., and Komatsu S. (2011) Changes in the plant proteome resulting from salt stress: Toward the creation of salt-tolerant crops? J. Proteomics 74, 1323–1337 - PubMed
-
- Zhu J. K. (2011) Plant salt tolerance. Trends Plant Sci. 6, 66–71 - PubMed
-
- Qu L. Q., Huang Y. Y., Zhu C. M., Zeng H. Q., Shen C. J., Liu C., Zhao Y., and Pi E. X. (2016) Rhizobia-inoculation enhances the soybean's tolerance to salt stress. Plant Soil 400, 209–222
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

