Distinct functions for the membrane-proximal ectodomain region (MPER) of HIV-1 gp41 in cell-free and cell-cell viral transmission and cell-cell fusion
- PMID: 29496992
- PMCID: PMC5912456
- DOI: 10.1074/jbc.RA117.000537
Distinct functions for the membrane-proximal ectodomain region (MPER) of HIV-1 gp41 in cell-free and cell-cell viral transmission and cell-cell fusion
Abstract
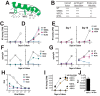
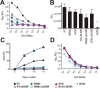
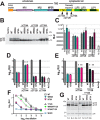
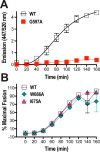
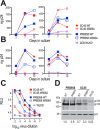
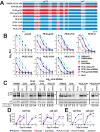
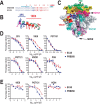
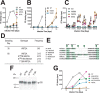
HIV-1 is spread by cell-free virions and by cell-cell viral transfer. We asked whether the structure and function of a broad neutralizing antibody (bNAb) epitope, the membrane-proximal ectodomain region (MPER) of the viral gp41 transmembrane glycoprotein, differ in cell-free and cell-cell-transmitted viruses and whether this difference could be related to Ab neutralization sensitivity. Whereas cell-free viruses bearing W666A and I675A substitutions in the MPER lacked infectivity, cell-associated mutant viruses were able to initiate robust spreading infection. Infectivity was restored to cell-free viruses by additional substitutions in the cytoplasmic tail (CT) of gp41 known to disrupt interactions with the viral matrix protein. We observed contrasting effects on cell-free virus infectivity when W666A was introduced to two transmitted/founder isolates, but both mutants could still mediate cell-cell spread. Domain swapping indicated that the disparate W666A phenotypes of the cell-free transmitted/founder viruses are controlled by sequences in variable regions 1, 2, and 4 of gp120. The sequential passaging of an MPER mutant (W672A) in peripheral blood mononuclear cells enabled selection of viral revertants with loss-of-glycan suppressor mutations in variable region 1, suggesting a functional interaction between variable region 1 and the MPER. An MPER-directed bNAb neutralized cell-free virus but not cell-cell viral spread. Our results suggest that the MPER of cell-cell-transmitted virions has a malleable structure that tolerates mutagenic disruption but is not accessible to bNAbs. In cell-free virions, interactions mediated by the CT impose an alternative MPER structure that is less tolerant of mutagenic alteration and is efficiently targeted by bNAbs.
Keywords: cell–cell transmission; glycoprotein; human immunodeficiency virus (HIV); membrane fusion; mutant; neutralization; viral replication; virus entry.
© 2018 by The American Society for Biochemistry and Molecular Biology, Inc.
Conflict of interest statement
The authors declare that they have no conflicts of interest with the contents of this article
Figures
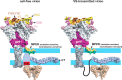
References
-
- Muranyi W., Malkusch S., Müller B., Heilemann M., and Kräusslich H. G. (2013) Super-resolution microscopy reveals specific recruitment of HIV-1 envelope proteins to viral assembly sites dependent on the envelope C-terminal tail. PLoS Pathog. 9, e1003198 10.1371/journal.ppat.1003198 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

