Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera
- PMID: 29497402
- PMCID: PMC5819568
- DOI: 10.3389/fmicb.2018.00067
Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera
Erratum in
-
Corrigendum: Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera.Front Microbiol. 2019 Apr 9;10:714. doi: 10.3389/fmicb.2019.00714. eCollection 2019. Front Microbiol. 2019. PMID: 31024493 Free PMC article.
Abstract
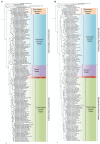
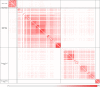
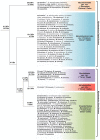
The genus Mycobacterium contains 188 species including several major human pathogens as well as numerous other environmental species. We report here comprehensive phylogenomics and comparative genomic analyses on 150 genomes of Mycobacterium species to understand their interrelationships. Phylogenetic trees were constructed for the 150 species based on 1941 core proteins for the genus Mycobacterium, 136 core proteins for the phylum Actinobacteria and 8 other conserved proteins. Additionally, the overall genome similarity amongst the Mycobacterium species was determined based on average amino acid identity of the conserved protein families. The results from these analyses consistently support the existence of five distinct monophyletic groups within the genus Mycobacterium at the highest level, which are designated as the "Tuberculosis-Simiae," "Terrae," "Triviale," "Fortuitum-Vaccae," and "Abscessus-Chelonae" clades. Some of these clades have also been observed in earlier phylogenetic studies. Of these clades, the "Abscessus-Chelonae" clade forms the deepest branching lineage and does not form a monophyletic grouping with the "Fortuitum-Vaccae" clade of fast-growing species. In parallel, our comparative analyses of proteins from mycobacterial genomes have identified 172 molecular signatures in the form of conserved signature indels and conserved signature proteins, which are uniquely shared by either all Mycobacterium species or by members of the five identified clades. The identified molecular signatures (or synapomorphies) provide strong independent evidence for the monophyly of the genus Mycobacterium and the five described clades and they provide reliable means for the demarcation of these clades and for their diagnostics. Based on the results of our comprehensive phylogenomic analyses and numerous identified molecular signatures, which consistently and strongly support the division of known mycobacterial species into the five described clades, we propose here division of the genus Mycobacterium into an emended genus Mycobacterium encompassing the "Tuberculosis-Simiae" clade, which includes all of the major human pathogens, and four novel genera viz. Mycolicibacterium gen. nov., Mycolicibacter gen. nov., Mycolicibacillus gen. nov. and Mycobacteroides gen. nov. corresponding to the "Fortuitum-Vaccae," "Terrae," "Triviale," and "Abscessus-Chelonae" clades, respectively. With the division of mycobacterial species into these five distinct groups, attention can now be focused on unique genetic and molecular characteristics that differentiate members of these groups.
Keywords: Mycobacterium classification; abscessus-chelonae clade; conserved signature indels and signature proteins; fortuitum-vaccae clade; phylogenomic analysis; slow-growing and fast-growing mycobacteria; terrae clade; triviale clade.
Figures







Comment in
-
Commentary: Phylogenomics and Comparative Genomic Studies Robustly Support Division of the Genus Mycobacterium into an Emended Genus Mycobacterium and Four Novel Genera.Front Microbiol. 2018 Sep 6;9:2065. doi: 10.3389/fmicb.2018.02065. eCollection 2018. Front Microbiol. 2018. PMID: 30237789 Free PMC article. No abstract available.
References
-
- Adékambi T., Berger P., Raoult D., Drancourt M. (2006a). rpoB gene sequence-based characterization of emerging non-tuberculous mycobacteria with descriptions of Mycobacterium bolletii sp. nov., Mycobacterium phocaicum sp. nov. and Mycobacterium aubagnense sp. nov. Int. J. Syst. Evol. Microbiol. 56, 133–143. 10.1099/ijs.0.63969-0 - DOI - PubMed
-
- Adékambi T., Stein A., Carvajal J., Raoult D., Drancourt M. (2006b). Description of Mycobacterium conceptionense sp. nov., a Mycobacterium fortuitum group organism isolated from a posttraumatic osteitis inflammation. J. Clin. Microbiol. 44, 1268–1273. 10.1128/JCM.44.4.1268-1273.2006 - DOI - PMC - PubMed
-
- Adeolu M., Alnajar S., Naushad S., Gupta S. (2016). Genome-based phylogeny and taxonomy of the ‘Enterobacteriales’: proposal for Enterobacterales ord. nov. divided into the families Enterobacteriaceae, Erwiniaceae fam. nov., Pectobacteriaceae fam. nov., Yersiniaceae fam. nov., Hafniaceae fam. nov., Morganellaceae fam. nov., and Budviciaceae fam. nov. Int. J. Syst. Evol. Microbiol. 66, 5575–5599. 10.1099/ijsem.0.001485 - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

