Protein cage assembly across multiple length scales
- PMID: 29497713
- PMCID: PMC6729141
- DOI: 10.1039/c7cs00818j
Protein cage assembly across multiple length scales
Abstract
Within the materials science community, proteins with cage-like architectures are being developed as versatile nanoscale platforms for use in protein nanotechnology. Much effort has been focused on the functionalization of protein cages with biological and non-biological moieties to bring about new properties of not only individual protein cages, but collective bulk-scale assemblies of protein cages. In this review, we report on the current understanding of protein cage assembly, both of the cages themselves from individual subunits, and the assembly of the individual protein cages into higher order structures. We start by discussing the key properties of natural protein cages (for example: size, shape and structure) followed by a review of some of the mechanisms of protein cage assembly and the factors that influence it. We then explore the current approaches for functionalizing protein cages, on the interior or exterior surfaces of the capsids. Lastly, we explore the emerging area of higher order assemblies created from individual protein cages and their potential for new and exciting collective properties.
Conflict of interest statement
Conflicts of interest
There are no conflicts of interest to declare.
Figures



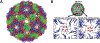

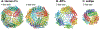

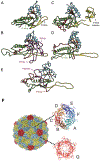

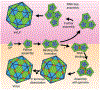
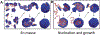
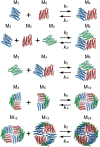
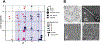
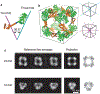

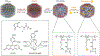
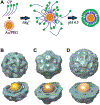




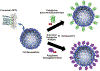

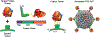



References
-
- Johnson JE and Chiu W, Curr. Opin. Struct. Biol, 2000, 10, 229–235. - PubMed
-
- Douglas T and Young M, Science, 2006, 312, 873–875. - PubMed
-
- Harrison PM and Arosio P, Biochim. Biophys. Acta, Bioenerg, 1996, 1275, 161–203. - PubMed
-
- Horwich AL, Fenton WA, Chapman E and Farr GW, Annu. Rev. Cell Dev. Biol, 2007, 23, 115–145. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

