Population-specific design of de-immunized protein biotherapeutics
- PMID: 29499035
- PMCID: PMC5851651
- DOI: 10.1371/journal.pcbi.1005983
Population-specific design of de-immunized protein biotherapeutics
Abstract
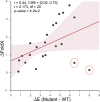
Immunogenicity is a major problem during the development of biotherapeutics since it can lead to rapid clearance of the drug and adverse reactions. The challenge for biotherapeutic design is therefore to identify mutants of the protein sequence that minimize immunogenicity in a target population whilst retaining pharmaceutical activity and protein function. Current approaches are moderately successful in designing sequences with reduced immunogenicity, but do not account for the varying frequencies of different human leucocyte antigen alleles in a specific population and in addition, since many designs are non-functional, require costly experimental post-screening. Here, we report a new method for de-immunization design using multi-objective combinatorial optimization. The method simultaneously optimizes the likelihood of a functional protein sequence at the same time as minimizing its immunogenicity tailored to a target population. We bypass the need for three-dimensional protein structure or molecular simulations to identify functional designs by automatically generating sequences using probabilistic models that have been used previously for mutation effect prediction and structure prediction. As proof-of-principle we designed sequences of the C2 domain of Factor VIII and tested them experimentally, resulting in a good correlation with the predicted immunogenicity of our model.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




References
-
- Leader B, Baca QJ, Golan DE. Protein therapeutics: a summary and pharmacological classification. Nature Reviews Drug Discovery. 2008;7(1):21–39. doi: 10.1038/nrd2399 - DOI - PubMed
-
- Jiang K. Near-record number of approvals signals drug development shift. Nature medicine. 2013;19(2):114–. doi: 10.1038/nm0213-114 - DOI - PubMed
-
- Schellekens H. Bioequivalence and the immunogenicity of biopharmaceuticals. Nature reviews Drug discovery. 2002;1(6):457–62. doi: 10.1038/nrd818 - DOI - PubMed
-
- Krieckaert C, Rispens T, Wolbink G. Immunogenicity of biological therapeutics: from assay to patient. Current opinion in rheumatology. 2012;24(3):306–11. doi: 10.1097/BOR.0b013e3283521c4e - DOI - PubMed
-
- Ryu JK, Kim HS, Nam DH. Current status and perspectives of biopharmaceutical drugs. Biotechnology and Bioprocess Engineering. 2012;17(5):900–11.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

