SETD7 Drives Cardiac Lineage Commitment through Stage-Specific Transcriptional Activation
- PMID: 29499155
- PMCID: PMC5929163
- DOI: 10.1016/j.stem.2018.02.005
SETD7 Drives Cardiac Lineage Commitment through Stage-Specific Transcriptional Activation
Abstract
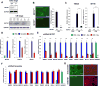
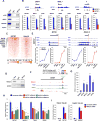
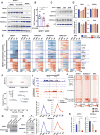
Cardiac development requires coordinated and large-scale rearrangements of the epigenome. The roles and precise mechanisms through which specific epigenetic modifying enzymes control cardiac lineage specification, however, remain unclear. Here we show that the H3K4 methyltransferase SETD7 controls cardiac differentiation by reading H3K36 marks independently of its enzymatic activity. Through chromatin immunoprecipitation sequencing (ChIP-seq), we found that SETD7 targets distinct sets of genes to drive their stage-specific expression during cardiomyocyte differentiation. SETD7 associates with different co-factors at these stages, including SWI/SNF chromatin-remodeling factors during mesodermal formation and the transcription factor NKX2.5 in cardiac progenitors to drive their differentiation. Further analyses revealed that SETD7 binds methylated H3K36 in the bodies of its target genes to facilitate RNA polymerase II (Pol II)-dependent transcription. Moreover, abnormal SETD7 expression impairs functional attributes of terminally differentiated cardiomyocytes. Together, these results reveal how SETD7 acts at sequential steps in cardiac lineage commitment, and they provide insights into crosstalk between dynamic epigenetic marks and chromatin-modifying enzymes.
Keywords: H3K36 methylation; SETD7; cardiomyocyte; epigenetics; histone modification; lineage commitment; stem cell; transcriptional regulation.
Copyright © 2018 Elsevier Inc. All rights reserved.
Figures




Comment in
-
SETD7 interacts with other chromatin-modifying factors to regulate cardiac development.Stem Cell Investig. 2019 Jun 27;6:14. doi: 10.21037/sci.2019.05.03. eCollection 2019. Stem Cell Investig. 2019. PMID: 31304180 Free PMC article. No abstract available.
-
SETD7 at the heart of chromatin factor interplay.Stem Cell Investig. 2019 Aug 1;6:20. doi: 10.21037/sci.2019.06.01. eCollection 2019. Stem Cell Investig. 2019. PMID: 31559307 Free PMC article. No abstract available.
-
Functions of SETD7 during development, homeostasis and cancer.Stem Cell Investig. 2019 Sep 2;6:26. doi: 10.21037/sci.2019.06.10. eCollection 2019. Stem Cell Investig. 2019. PMID: 31620473 Free PMC article. No abstract available.
-
SETD7 in cardiomyocyte differentiation and cardiac function.Stem Cell Investig. 2019 Sep 9;6:29. doi: 10.21037/sci.2019.08.01. eCollection 2019. Stem Cell Investig. 2019. PMID: 31620476 Free PMC article. No abstract available.
References
-
- Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial Distribution of Di- and Tri-methyl Lysine 36 of Histone H3 at Active Genes. J. Biol. Chem. 2005;280:17732–17736. - PubMed
-
- Brien GL, Gambero G, O’Connell DJ, Jerman E, Turner SA, Egan CM, Dunne EJ, Jurgens MC, Wynne K, Piao L, et al. Polycomb PHF19 binds H3K36me3 and recruits PRC2 and demethylase NO66 to embryonic stem cell genes during differentiation. Nat. Struct. Mol. Biol. 2012;19:1273–1281. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

