Discovery and Biosynthesis of the Antibiotic Bicyclomycin in Distantly Related Bacterial Classes
- PMID: 29500259
- PMCID: PMC5930311
- DOI: 10.1128/AEM.02828-17
Discovery and Biosynthesis of the Antibiotic Bicyclomycin in Distantly Related Bacterial Classes
Abstract
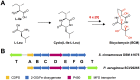
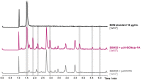
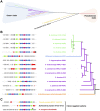
Bicyclomycin (BCM) is a clinically promising antibiotic that is biosynthesized by Streptomyces cinnamoneus DSM 41675. BCM is structurally characterized by a core cyclo(l-Ile-l-Leu) 2,5-diketopiperazine (DKP) that is extensively oxidized. Here, we identify the BCM biosynthetic gene cluster, which shows that the core of BCM is biosynthesized by a cyclodipeptide synthase, and the oxidative modifications are introduced by five 2-oxoglutarate-dependent dioxygenases and one cytochrome P450 monooxygenase. The discovery of the gene cluster enabled the identification of BCM pathways encoded by the genomes of hundreds of Pseudomonas aeruginosa isolates distributed globally, and heterologous expression of the pathway from P. aeruginosa SCV20265 demonstrated that the product is chemically identical to BCM produced by S. cinnamoneus Overall, putative BCM gene clusters have been found in at least seven genera spanning Actinobacteria and Proteobacteria (Alphaproteobacteria, Betaproteobacteria, and Gammaproteobacteria). This represents a rare example of horizontal gene transfer of an intact biosynthetic gene cluster across such distantly related bacteria, and we show that these gene clusters are almost always associated with mobile genetic elements.IMPORTANCE Bicyclomycin is the only natural product antibiotic that selectively inhibits the transcription termination factor Rho. This mechanism of action, combined with its proven biological safety and its activity against clinically relevant Gram-negative bacterial pathogens, makes it a very promising antibiotic candidate. Here, we report the identification of the bicyclomycin biosynthetic gene cluster in the known bicyclomycin-producing organism Streptomyces cinnamoneus, which will enable the engineered production of new bicyclomycin derivatives. The identification of this gene cluster also led to the discovery of hundreds of bicyclomycin pathways encoded in highly diverse bacteria, including in the opportunistic pathogen Pseudomonas aeruginosa This wide distribution of a complex biosynthetic pathway is very unusual and provides an insight into how a pathway for an antibiotic can be transferred between diverse bacteria.
Keywords: Pseudomonas aeruginosa; Streptomyces; antibiotic; biosynthesis; gene transfer; phylogenetic analysis.
Copyright © 2018 Vior et al.
Figures

Similar articles
-
Study of bicyclomycin biosynthesis in Streptomyces cinnamoneus by genetic and biochemical approaches.Sci Rep. 2019 Dec 27;9(1):20226. doi: 10.1038/s41598-019-56747-7. Sci Rep. 2019. PMID: 31882990 Free PMC article.
-
Identification of the Biosynthetic Pathway for the Antibiotic Bicyclomycin.Biochemistry. 2018 Jan 9;57(1):61-65. doi: 10.1021/acs.biochem.7b00943. Epub 2017 Nov 7. Biochemistry. 2018. PMID: 29053243 Free PMC article.
-
A Six-Oxidase Cascade for Tandem C-H Bond Activation Revealed by Reconstitution of Bicyclomycin Biosynthesis.Angew Chem Int Ed Engl. 2018 Jan 15;57(3):719-723. doi: 10.1002/anie.201710529. Epub 2017 Dec 12. Angew Chem Int Ed Engl. 2018. PMID: 29194897
-
The molecular basis for the mode of action of bicyclomycin.Curr Drug Targets Infect Disord. 2005 Sep;5(3):273-95. doi: 10.2174/1568005054880136. Curr Drug Targets Infect Disord. 2005. PMID: 16181146 Review.
-
Polyene antibiotic biosynthesis gene clusters.Appl Microbiol Biotechnol. 2003 May;61(3):179-88. doi: 10.1007/s00253-002-1183-5. Epub 2002 Dec 18. Appl Microbiol Biotechnol. 2003. PMID: 12698274 Review.
Cited by
-
The Desotamide Family of Antibiotics.Antibiotics (Basel). 2020 Jul 27;9(8):452. doi: 10.3390/antibiotics9080452. Antibiotics (Basel). 2020. PMID: 32727132 Free PMC article. Review.
-
Critical analysis of polycyclic tetramate macrolactam biosynthetic gene cluster phylogeny and functional diversity.Appl Environ Microbiol. 2024 Jun 18;90(6):e0060024. doi: 10.1128/aem.00600-24. Epub 2024 May 21. Appl Environ Microbiol. 2024. PMID: 38771054 Free PMC article.
-
Identification of the bacterial metabolite aerugine as potential trigger of human dopaminergic neurodegeneration.Environ Int. 2023 Oct;180:108229. doi: 10.1016/j.envint.2023.108229. Epub 2023 Sep 23. Environ Int. 2023. PMID: 37797477 Free PMC article.
-
Crystal Structure and Mutagenesis of an XYP Subfamily Cyclodipeptide Synthase Reveal Key Determinants of Enzyme Activity and Substrate Specificity.Biochemistry. 2024 Nov 19;63(22):2969-2976. doi: 10.1021/acs.biochem.4c00505. Epub 2024 Oct 30. Biochemistry. 2024. PMID: 39475147 Free PMC article.
-
Beyond Soil-Dwelling Actinobacteria: Fantastic Antibiotics and Where to Find Them.Antibiotics (Basel). 2022 Feb 2;11(2):195. doi: 10.3390/antibiotics11020195. Antibiotics (Basel). 2022. PMID: 35203798 Free PMC article. Review.
References
-
- Miyoshi T, Miyairi N, Aoki H, Kohsaka M, Sakai H. 1972. Bicyclomycin, a new antibiotic. I. Taxonomy, isolation and characterization J Antibiot (Tokyo) 25:569–575. - PubMed
-
- Kamiya T, Maeno S, Hashimoto M, Mine Y. 1972. Bicyclomycin, a new antibiotic. II. Structural elucidation and acyl derivatives. J Antibiot (Tokyo) 25:576–581. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- BBS/E/J/000PR9790/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/P012523/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- MR/P007570/1/MRC_/Medical Research Council/United Kingdom
- BB/J004561/1 /BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials

