Improved isolation strategies to increase the yield and purity of human urinary exosomes for biomarker discovery
- PMID: 29500443
- PMCID: PMC5834546
- DOI: 10.1038/s41598-018-22142-x
Improved isolation strategies to increase the yield and purity of human urinary exosomes for biomarker discovery
Abstract
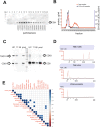
Circulating miRNAs are detected in extracellular space and body fluids such as urine. Circulating RNAs can be packaged in secreted urinary extracellular vesicles (uEVs) and thus protected from degradation. Urinary exosome preparations might contain specific miRNAs, relevant as biomarkers in renal and bladder diseases. Major difficulties in application of uEVs into the clinical environment are the high variability and low reproducibility of uEV isolation methods. Here we used five different methods to isolate uEVs and compared the size distribution, morphology, yield, presence of exosomal protein markers and RNA content of uEVs. We present an optimized ultracentrifugation and size exclusion chromatography approach for highly reproducible isolation for 50-150 nm uEVs, corresponding to the exosomes, from 50 ml urine. We profiled the miRNA content of uEVs and total urine from the same samples with the NanoString platform and validated the data using qPCR. Our results indicate that 18 miRNAs, robustly detected in uEVs were always present in the total urine. However, 15 miRNAs could be detected only in the total urine preparations and might represent naked circulating miRNA species. This is a novel unbiased and reproducible strategy for uEVs isolation, content normalization and miRNA cargo analysis, suitable for biomarker discovery studies.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
-
- Duan LJ, et al. miR-133 modulates TGF-beta 1-induced bladder smooth muscle cell hypertrophic and fibrotic response: Implication for a role of microRNA in bladder wall remodeling caused by bladder outlet obstruction. Cellular Signalling. 2015;27:215–227. doi: 10.1016/j.cellsig.2014.11.001. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

