Peptide identifications and false discovery rates using different mass spectrometry platforms
- PMID: 29501178
- PMCID: PMC5839655
- DOI: 10.1016/j.talanta.2018.01.062
Peptide identifications and false discovery rates using different mass spectrometry platforms
Abstract
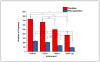
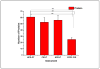
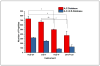
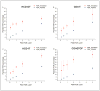
Characterization of endogenous neuropeptides produced from post-translational proteolytic processing of precursor proteins is a demanding task. A variety of complex prohormone processing steps generate molecular diversity from neuropeptide prohormones, making in silico neuropeptide discovery difficult. In addition, the wide range of endogenous peptide concentrations as well as significant peptide complexity further challenge the structural characterization of neuropeptides. Liquid chromatography-mass spectrometry (MS), performed in conjunction with bioinformatics, allows for high-throughput characterization of peptides. Mass analyzers and molecular dissociation techniques render specific characteristics to the acquired data and thus, influence the analysis of the MS data using bioinformatic algorithms for follow-up peptide identification. Here we evaluated the efficacy of several distinct peptidomic workflows using two mass spectrometers, the Thermo Orbitrap Fusion Tribrid and Bruker Impact HD UHR-QqTOF, for confident peptide discovery and characterization. We compared the results in several categories, including the numbers of identified peptides, full-length mature neuropeptides among all identifications, and precursor proteins mapped by the identified peptides. We also characterized the peptide false discovery rate (FDR) based on the occurrence of amidation, a known post-translational modification (PTM) that has been shown to require the presence of a C-terminal glycine. Thus, amidation events without a preceding glycine were considered false-positive amidation assignments. We compared the FDR calculated by the search engine used here to the minimum FDR estimated via false amidation assignments. The search engine severely underestimated the rate of false PTM assignments among the identified peptides, regardless of the specific MS platform used.
Keywords: Amidation; De novo sequencing; Neuropeptides; Peptidomics; Proteomics.
Copyright © 2018 Elsevier B.V. All rights reserved.
Figures

Similar articles
-
Assessment and Comparison of Database Search Engines for Peptidomic Applications.J Proteome Res. 2023 Oct 6;22(10):3123-3134. doi: 10.1021/acs.jproteome.2c00307. Epub 2023 Feb 21. J Proteome Res. 2023. PMID: 36809008 Free PMC article.
-
Improving the identification rate of endogenous peptides using electron transfer dissociation and collision-induced dissociation.J Proteome Res. 2013 Dec 6;12(12):5410-21. doi: 10.1021/pr400446z. Epub 2013 Oct 3. J Proteome Res. 2013. PMID: 24032530
-
Diversity of Neuropeptide Cell-Cell Signaling Molecules Generated by Proteolytic Processing Revealed by Neuropeptidomics Mass Spectrometry.J Am Soc Mass Spectrom. 2018 May;29(5):807-816. doi: 10.1007/s13361-018-1914-1. Epub 2018 Apr 17. J Am Soc Mass Spectrom. 2018. PMID: 29667161 Free PMC article. Review.
-
Peptidomics of the zebrafish Danio rerio: In search for neuropeptides.J Proteomics. 2017 Jan 6;150:290-296. doi: 10.1016/j.jprot.2016.09.015. Epub 2016 Oct 2. J Proteomics. 2017. PMID: 27705817
-
Modification Site Localization in Peptides.Adv Exp Med Biol. 2016;919:243-247. doi: 10.1007/978-3-319-41448-5_13. Adv Exp Med Biol. 2016. PMID: 27975222 Review.
Cited by
-
PACAP and Other Neuropeptide Targets Link Chronic Migraine and Opioid-induced Hyperalgesia in Mouse Models.Mol Cell Proteomics. 2019 Dec;18(12):2447-2458. doi: 10.1074/mcp.RA119.001767. Epub 2019 Oct 24. Mol Cell Proteomics. 2019. PMID: 31649062 Free PMC article.
-
Quantitative neuropeptide analysis by mass spectrometry: advancing methodologies for biological discovery.RSC Chem Biol. 2025 Jun 12;6(8):1214-1232. doi: 10.1039/d5cb00082c. eCollection 2025 Jul 30. RSC Chem Biol. 2025. PMID: 40529571 Free PMC article. Review.
-
Proteomics and machine learning: Leveraging domain knowledge for feature selection in a skeletal muscle tissue meta-analysis.Heliyon. 2024 Nov 29;10(24):e40772. doi: 10.1016/j.heliyon.2024.e40772. eCollection 2024 Dec 30. Heliyon. 2024. PMID: 39720035 Free PMC article.
-
Neuropeptidomics of the American Lobster Homarus americanus.J Proteome Res. 2024 May 3;23(5):1757-1767. doi: 10.1021/acs.jproteome.3c00925. Epub 2024 Apr 22. J Proteome Res. 2024. PMID: 38644788 Free PMC article.
-
Recent advances in mass spectrometry analysis of neuropeptides.Mass Spectrom Rev. 2023 Mar;42(2):706-750. doi: 10.1002/mas.21734. Epub 2021 Sep 24. Mass Spectrom Rev. 2023. PMID: 34558119 Free PMC article. Review.
References
-
- Arora S, Anubhuti Role of neuropeptides in appetite regulation and obesity–A review. Neuropeptides. 2006;40:375–401. - PubMed
-
- Trayhurn P, Beattie JH. Physiological role of adipose tissue: white adipose tissue as an endocrine and secretory organ. Proc Nutr Soc. 2001;60:329–339. - PubMed
-
- De Souza EB. Corticotropin-releasing factor receptors: Physiology, pharmacology, biochemistry and role in central nervous system and immune disorders. Psychoneuroendocrinology. 1995;20:789–819. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

